Quantification of Cable Bacteria in Marine Sediments via qPCR
- PMID: 32719667
- PMCID: PMC7348212
- DOI: 10.3389/fmicb.2020.01506
Quantification of Cable Bacteria in Marine Sediments via qPCR
Abstract
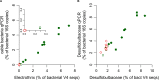
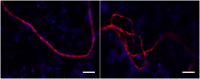
Cable bacteria (Deltaproteobacteria, Desulfobulbaceae) are long filamentous sulfur-oxidizing bacteria that generate long-distance electric currents running through the bacterial filaments. This way, they couple the oxidation of sulfide in deeper sediment layers to the reduction of oxygen or nitrate near the sediment-water interface. Cable bacteria are found in a wide range of aquatic sediments, but an accurate procedure to assess their abundance is lacking. We developed a qPCR approach that quantifies cable bacteria in relation to other bacteria within the family Desulfobulbaceae. Primer sets targeting cable bacteria, Desulfobulbaceae and the total bacterial community were applied in qPCR with DNA extracted from marine sediment incubations. Amplicon sequencing of the 16S rRNA gene V4 region confirmed that cable bacteria were accurately enumerated by qPCR, and suggested novel diversity of cable bacteria. The conjoint quantification of current densities and cell densities revealed that individual filaments carry a mean current of ∼110 pA and have a cell specific oxygen consumption rate of 69 fmol O2 cell-1 day-1. Overall, the qPCR method enables a better quantitative assessment of cable bacteria abundance, providing new metabolic insights at filament and cell level, and improving our understanding of the microbial ecology of electrogenic sediments.
Keywords: Desulfobulbaceae; amplicon sequencing; cable bacteria; current density; marine sediment; oxygen consumption rate; quantitative PCR.
Copyright © 2020 Geelhoed, van de Velde and Meysman.
Figures



Similar articles
-
Identification of cable bacteria and its biogeochemical impact on sulfur in freshwater sediments from the Wenyu River.Sci Total Environ. 2021 May 15;769:144541. doi: 10.1016/j.scitotenv.2020.144541. Epub 2021 Jan 14. Sci Total Environ. 2021. PMID: 33482557
-
Cable Bacteria in Freshwater Sediments.Appl Environ Microbiol. 2015 Sep 1;81(17):6003-11. doi: 10.1128/AEM.01064-15. Epub 2015 Jun 26. Appl Environ Microbiol. 2015. PMID: 26116678 Free PMC article.
-
Microbial carbon metabolism associated with electrogenic sulphur oxidation in coastal sediments.ISME J. 2015 Sep;9(9):1966-78. doi: 10.1038/ismej.2015.10. Epub 2015 Feb 13. ISME J. 2015. PMID: 25679534 Free PMC article.
-
Electrogenic sulfur oxidation mediated by cable bacteria and its ecological effects.Environ Sci Ecotechnol. 2023 Dec 19;20:100371. doi: 10.1016/j.ese.2023.100371. eCollection 2024 Jul. Environ Sci Ecotechnol. 2023. PMID: 38283867 Free PMC article. Review.
-
Rethinking sediment biogeochemistry after the discovery of electric currents.Ann Rev Mar Sci. 2015;7:425-42. doi: 10.1146/annurev-marine-010814-015708. Epub 2014 Sep 19. Ann Rev Mar Sci. 2015. PMID: 25251266 Review.
Cited by
-
Long-distance electron transport in multicellular freshwater cable bacteria.Elife. 2024 Aug 29;12:RP91097. doi: 10.7554/eLife.91097. Elife. 2024. PMID: 39207443 Free PMC article.
-
Indications for a genetic basis for big bacteria and description of the giant cable bacterium Candidatus Electrothrix gigas sp. nov.Microbiol Spectr. 2023 Sep 21;11(5):e0053823. doi: 10.1128/spectrum.00538-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37732806 Free PMC article.
-
Protocol for using autoclaved intertidal sediment as a medium to enrich marine cable bacteria.STAR Protoc. 2022 Aug 11;3(3):101604. doi: 10.1016/j.xpro.2022.101604. eCollection 2022 Sep 16. STAR Protoc. 2022. PMID: 35990745 Free PMC article.
-
Using Oxidative Electrodes to Enrich Novel Members in the Desulfobulbaceae Family from Intertidal Sediments.Microorganisms. 2021 Nov 11;9(11):2329. doi: 10.3390/microorganisms9112329. Microorganisms. 2021. PMID: 34835454 Free PMC article.
-
Comparative genomic analysis of nickel homeostasis in cable bacteria.BMC Genomics. 2024 Jul 15;25(1):692. doi: 10.1186/s12864-024-10594-7. BMC Genomics. 2024. PMID: 39009997 Free PMC article.
References
-
- Agogue H., Brink M., Dinasquet J., Herndl G. J. (2008). Major gradients in putatively nitrifying and non-nitrifying Archaea in the deep North Atlantic. Nature 456 788–U772. - PubMed
-
- Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A. (1990). Combination of 16S ribosomal-RNA-targeted oligonucleotide probes with flow-cytometry for analyzing mixed microbial-populations. Appl. Environ. Microbiol. 56 1919–1925. 10.1128/aem.56.6.1919-1925.1990 - DOI - PMC - PubMed
-
- Burdorf L. D. W., Hidalgo-Martinez S., Cook P. L. M., Meysman F. J. R. (2016). Long-distance electron transport by cable bacteria in mangrove sediments. Mar. Ecol. Progr. Ser. 545 1–8. 10.3354/meps11635 - DOI
LinkOut - more resources
Full Text Sources

