Histone lysine demethylase 3B (KDM3B) regulates the propagation of autophagy via transcriptional activation of autophagy-related genes
- PMID: 32716961
- PMCID: PMC7384621
- DOI: 10.1371/journal.pone.0236403
Histone lysine demethylase 3B (KDM3B) regulates the propagation of autophagy via transcriptional activation of autophagy-related genes
Abstract
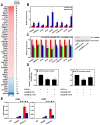
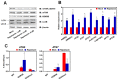
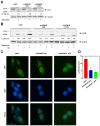
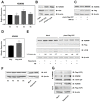
Autophagy, a self-degradative physiological process, is critical for homeostasis maintenance and energy source balancing in response to various stresses, including nutrient deprivation. It is a highly conserved catabolic process in eukaryotes and is indispensable for cell survival as it involves degradation of unessential or excessive components and their subsequent recycling as building blocks for the synthesis of necessary molecules. Although the dysregulation of autophagy has been reported to broadly contribute to various diseases, including cancers and neurodegenerative diseases, the molecular mechanisms underlying the epigenetic regulation of autophagy are poorly elucidated. Here, we report that the level of lysine demethylase 3B (KDM3B) increases in nutrient-deprived HCT116 cells, a colorectal carcinoma cell line, resulting in transcriptional activation of the autophagy-inducing genes. KDM3B was found to enhance the transcription by demethylating H3K9me2 on the promoter of these genes. Furthermore, we observed that the depletion of KDM3B inhibited the autophagic flux in HCT116 cells. Collectively, these data suggested the critical role of KDM3B in the regulation of autophagy-related genes via H3K9me2 demethylation and induction of autophagy in nutrient-starved HCT116 cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Histone lysine demethylase 3B regulates autophagy via transcriptional regulation of GABARAPL1 in acute myeloid leukemia cells.Int J Oncol. 2023 Jul;63(1):87. doi: 10.3892/ijo.2023.5535. Epub 2023 Jun 16. Int J Oncol. 2023. PMID: 37326062 Free PMC article.
-
Transcriptional repression of ANGPT1 by histone H3K9 demethylase KDM3B.BMB Rep. 2015 Jul;48(7):401-6. doi: 10.5483/bmbrep.2015.48.7.188. BMB Rep. 2015. PMID: 25413303 Free PMC article.
-
Histone demethylase KDM3B protects against ferroptosis by upregulating SLC7A11.FEBS Open Bio. 2020 Apr;10(4):637-643. doi: 10.1002/2211-5463.12823. Epub 2020 Mar 18. FEBS Open Bio. 2020. PMID: 32107878 Free PMC article.
-
Epigenetic regulation of autophagy by histone-modifying enzymes under nutrient stress.Cell Death Differ. 2023 Jun;30(6):1430-1436. doi: 10.1038/s41418-023-01154-9. Epub 2023 Mar 30. Cell Death Differ. 2023. PMID: 36997734 Free PMC article. Review.
-
The future therapeutic potential of histone demethylases: A critical analysis.Curr Opin Drug Discov Devel. 2009 Sep;12(5):607-15. Curr Opin Drug Discov Devel. 2009. PMID: 19736620 Review.
Cited by
-
Kdm3b haploinsufficiency impairs the consolidation of cerebellum-dependent motor memory in mice.Mol Brain. 2021 Jul 3;14(1):106. doi: 10.1186/s13041-021-00815-5. Mol Brain. 2021. PMID: 34217333 Free PMC article.
-
Histone lysine demethylase 3B regulates autophagy via transcriptional regulation of GABARAPL1 in acute myeloid leukemia cells.Int J Oncol. 2023 Jul;63(1):87. doi: 10.3892/ijo.2023.5535. Epub 2023 Jun 16. Int J Oncol. 2023. PMID: 37326062 Free PMC article.
-
Epigenetic regulation of autophagy-related genes: Implications for neurodevelopmental disorders.Autophagy. 2024 Jan;20(1):15-28. doi: 10.1080/15548627.2023.2250217. Epub 2023 Sep 6. Autophagy. 2024. PMID: 37674294 Free PMC article. Review.
References
-
- Mizushima N. Autophagy: process and function. Genes & development. 2007;21(22):2861–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

