Fungi, bacteria and oomycota opportunistically isolated from the seagrass, Zostera marina
- PMID: 32697800
- PMCID: PMC7375540
- DOI: 10.1371/journal.pone.0236135
Fungi, bacteria and oomycota opportunistically isolated from the seagrass, Zostera marina
Erratum in
-
Correction: Fungi, bacteria and oomycota opportunistically isolated from the seagrass, Zostera marina.PLoS One. 2021 May 6;16(5):e0251536. doi: 10.1371/journal.pone.0251536. eCollection 2021. PLoS One. 2021. PMID: 33956899 Free PMC article.
Abstract
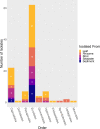
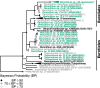
Fungi in the marine environment are often neglected as a research topic, despite that fungi having critical roles on land as decomposers, pathogens or endophytes. Here we used culture-dependent methods to survey the fungi associated with the seagrass, Zostera marina, also obtaining bacteria and oomycete isolates in the process. A total of 108 fungi, 40 bacteria and 2 oomycetes were isolated. These isolates were then taxonomically identified using a combination of molecular and phylogenetic methods. The majority of the fungal isolates were classified as belonging to the classes Eurotiomycetes, Dothideomycetes, and Sordariomycetes. Most fungal isolates were habitat generalists like Penicillium sp. and Cladosporium sp., but we also cultured a diverse set of rare taxa including possible habitat specialists like Colletotrichum sp. which may preferentially associate with Z. marina leaf tissue. Although the bulk of bacterial isolates were identified as being from known ubiquitous marine lineages, we also obtained several Actinomycetes isolates and a Phyllobacterium sp. We identified two oomycetes, another understudied group of marine microbial eukaryotes, as Halophytophthora sp. which may be opportunistic pathogens or saprophytes of Z. marina. Overall, this study generates a culture collection of fungi which adds to knowledge of Z. marina associated fungi and highlights a need for more investigation into the functional and evolutionary roles of microbial eukaryotes associated with seagrasses.
Conflict of interest statement
JAE is on the Scientific Advisory Board of Zymo Research, Inc. CLE declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures





Similar articles
-
Global Diversity and Biogeography of the Zostera marina Mycobiome.Appl Environ Microbiol. 2021 May 26;87(12):e0279520. doi: 10.1128/AEM.02795-20. Epub 2021 May 26. Appl Environ Microbiol. 2021. PMID: 33837008 Free PMC article.
-
Rapid Metabolome and Bioactivity Profiling of Fungi Associated with the Leaf and Rhizosphere of the Baltic Seagrass Zostera marina.Mar Drugs. 2019 Jul 19;17(7):419. doi: 10.3390/md17070419. Mar Drugs. 2019. PMID: 31330983 Free PMC article.
-
Characterization of the Mycobiome of the Seagrass, Zostera marina, Reveals Putative Associations With Marine Chytrids.Front Microbiol. 2019 Nov 1;10:2476. doi: 10.3389/fmicb.2019.02476. eCollection 2019. Front Microbiol. 2019. PMID: 31749781 Free PMC article.
-
Viruses of fungi and oomycetes in the soil environment.FEMS Microbiol Ecol. 2019 Sep 1;95(9):fiz119. doi: 10.1093/femsec/fiz119. FEMS Microbiol Ecol. 2019. PMID: 31365065 Review.
-
Interspecific hybridization in plant-associated fungi and oomycetes: a review.Mol Ecol. 2003 Nov;12(11):2861-73. doi: 10.1046/j.1365-294x.2003.01965.x. Mol Ecol. 2003. PMID: 14629368 Review.
Cited by
-
Global Diversity and Biogeography of the Zostera marina Mycobiome.Appl Environ Microbiol. 2021 May 26;87(12):e0279520. doi: 10.1128/AEM.02795-20. Epub 2021 May 26. Appl Environ Microbiol. 2021. PMID: 33837008 Free PMC article.
-
Multigene Phylogeny, Diversity and Antimicrobial Potential of Endophytic Sordariomycetes From Rosa roxburghii.Front Microbiol. 2021 Nov 29;12:755919. doi: 10.3389/fmicb.2021.755919. eCollection 2021. Front Microbiol. 2021. PMID: 34912312 Free PMC article.
-
Novel insights into the rhizosphere and seawater microbiome of Zostera marina in diverse mariculture zones.Microbiome. 2024 Feb 14;12(1):27. doi: 10.1186/s40168-024-01759-3. Microbiome. 2024. PMID: 38350953 Free PMC article.
-
Why Are There So Few Basidiomycota and Basal Fungi as Endophytes? A Review.J Fungi (Basel). 2024 Jan 15;10(1):67. doi: 10.3390/jof10010067. J Fungi (Basel). 2024. PMID: 38248976 Free PMC article. Review.
-
Fungal biodiversity and conservation mycology in light of new technology, big data, and changing attitudes.Curr Biol. 2021 Oct 11;31(19):R1312-R1325. doi: 10.1016/j.cub.2021.06.083. Curr Biol. 2021. PMID: 34637742 Free PMC article. Review.
References
-
- Jones EBG. Are there more marine fungi to be described? Botanica Marina. 2011. 10.1515/bot.2011.043 - DOI
-
- Jones EBG, Suetrong S, Sakayaroj J, Bahkali AH, Abdel-Wahab MA, Boekhout T, et al. Classification of marine Ascomycota, Basidiomycota, Blastocladiomycota and Chytridiomycota. Fungal Diversity. 2015. pp. 1–72. 10.1007/s13225-015-0339-4 - DOI
-
- Fourqurean JW, Duarte CM, Kennedy H, Marbà N, Holmer M, Mateo MA, et al. Seagrass ecosystems as a globally significant carbon stock. Nature Geoscience. 2012. pp. 505–509. 10.1038/ngeo1477 - DOI

