Determining Binding Kinetics of Intrinsically Disordered Proteins by NMR Spectroscopy
- PMID: 32696383
- PMCID: PMC7605514
- DOI: 10.1007/978-1-0716-0524-0_34
Determining Binding Kinetics of Intrinsically Disordered Proteins by NMR Spectroscopy
Abstract
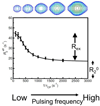
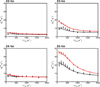
The unique structural flexibility of intrinsically disordered proteins (IDPs) is central to their diverse functions in cellular processes. Protein-protein interactions involving IDPs are frequently transient and dynamic in nature. Nuclear magnetic resonance (NMR) spectroscopy is an especially powerful tool for characterizing the structural propensities, dynamics, and interactions of IDPs. Here we describe applications of the Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion experiment in combination with NMR titrations to characterize the kinetics and mechanisms of interactions between intrinsically disordered proteins and their targets. We illustrate the method with reference to interactions between the activation domain of the human T-cell leukemia virus type-I (HTLV-1) basic leucine zipper protein (HBZ) and its cellular binding partner, the KIX domain of the transcriptional coactivator CBP.
Keywords: CPMG; IDP; Protein dynamics; Protein interaction; Relaxation dispersion.
Figures


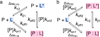
Similar articles
-
Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.Proc Natl Acad Sci U S A. 2018 Oct 2;115(40):10040-10045. doi: 10.1073/pnas.1810397115. Epub 2018 Sep 19. Proc Natl Acad Sci U S A. 2018. PMID: 30232260 Free PMC article.
-
Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding.Proc Natl Acad Sci U S A. 2015 Aug 4;112(31):9614-9. doi: 10.1073/pnas.1512799112. Epub 2015 Jul 20. Proc Natl Acad Sci U S A. 2015. PMID: 26195786 Free PMC article.
-
Deciphering the Dynamic Interaction Profile of an Intrinsically Disordered Protein by NMR Exchange Spectroscopy.J Am Chem Soc. 2018 Jan 24;140(3):1148-1158. doi: 10.1021/jacs.7b12407. Epub 2018 Jan 11. J Am Chem Soc. 2018. PMID: 29276882
-
NMR Spectroscopic Studies of the Conformational Ensembles of Intrinsically Disordered Proteins.Adv Exp Med Biol. 2015;870:149-85. doi: 10.1007/978-3-319-20164-1_5. Adv Exp Med Biol. 2015. PMID: 26387102 Review.
-
In-Cell NMR Spectroscopy of Intrinsically Disordered Proteins.Proteomics. 2019 Mar;19(6):e1800055. doi: 10.1002/pmic.201800055. Epub 2019 Jan 15. Proteomics. 2019. PMID: 30489014 Free PMC article. Review.
Cited by
-
NMR methods for exploring 'dark' states in ligand binding and protein-protein interactions.Prog Nucl Magn Reson Spectrosc. 2022 Feb;128:1-24. doi: 10.1016/j.pnmrs.2021.10.001. Epub 2021 Nov 2. Prog Nucl Magn Reson Spectrosc. 2022. PMID: 35282867 Free PMC article. Review.
-
NMR spectroscopy, excited states and relevance to problems in cell biology - transient pre-nucleation tetramerization of huntingtin and insights into Huntington's disease.J Cell Sci. 2022 Jun 15;135(12):jcs258695. doi: 10.1242/jcs.258695. Epub 2022 Jun 15. J Cell Sci. 2022. PMID: 35703323 Free PMC article.
References
-
- Loria JP, Rance M, Palmer AG (1999) A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J Am Chem Soc 121 (10):2331–2332. doi:DOI 10.1021/ja983961a - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

