Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome
- PMID: 32690954
- PMCID: PMC7508674
- DOI: 10.1038/s41564-020-0755-4
Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome
Abstract
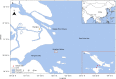
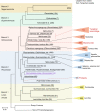
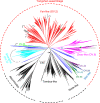
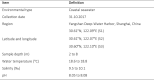
RNA viruses in aquatic environments remain poorly studied. Here, we analysed the RNA virome from approximately 10 l water from Yangshan Deep-Water Harbour near the Yangtze River estuary in China and identified more than 4,500 distinct RNA viruses, doubling the previously known set of viruses. Phylogenomic analysis identified several major lineages, roughly, at the taxonomic ranks of class, order and family. The 719-member-strong Yangshan virus assemblage is the sister clade to the expansive class Alsuviricetes and consists of viruses with simple genomes that typically encode only RNA-dependent RNA polymerase (RdRP), capping enzyme and capsid protein. Several clades within the Yangshan assemblage independently evolved domain permutation in the RdRP. Another previously unknown clade shares ancestry with Potyviridae, the largest known plant virus family. The 'Aquatic picorna-like viruses/Marnaviridae' clade was greatly expanded, with more than 800 added viruses. Several RdRP-linked protein domains not previously detected in any RNA viruses were identified, such as the small ubiquitin-like modifier (SUMO) domain, phospholipase A2 and PrsW-family protease domain. Multiple viruses utilize alternative genetic codes implying protist (especially ciliate) hosts. The results reveal a vast RNA virome that includes many previously unknown groups. However, phylogenetic analysis of the RdRPs supports the previously established five-branch structure of the RNA virus evolutionary tree, with no additional phyla.
Conflict of interest statement
The authors declare no competing interests.
Figures








Similar articles
-
Origins and Evolution of the Global RNA Virome.mBio. 2018 Nov 27;9(6):e02329-18. doi: 10.1128/mBio.02329-18. mBio. 2018. PMID: 30482837 Free PMC article.
-
Metagenome-Assembled Viral Genomes Analysis Reveals Diversity and Infectivity of the RNA Virome of Gerbillinae Species.Viruses. 2022 Feb 9;14(2):356. doi: 10.3390/v14020356. Viruses. 2022. PMID: 35215951 Free PMC article.
-
Metagenomics reshapes the concepts of RNA virus evolution by revealing extensive horizontal virus transfer.Virus Res. 2018 Jan 15;244:36-52. doi: 10.1016/j.virusres.2017.10.020. Epub 2017 Nov 8. Virus Res. 2018. PMID: 29103997 Free PMC article. Review.
-
Diversity of Picorna-Like Viruses in the Teltow Canal, Berlin, Germany.Viruses. 2024 Jun 25;16(7):1020. doi: 10.3390/v16071020. Viruses. 2024. PMID: 39066183 Free PMC article.
-
Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences.Crit Rev Biochem Mol Biol. 1993;28(5):375-430. doi: 10.3109/10409239309078440. Crit Rev Biochem Mol Biol. 1993. PMID: 8269709 Review.
Cited by
-
Freshwater Mussel Viromes Increase Rapidly in Diversity and Abundance When Hosts Are Released from Captivity into the Wild.Animals (Basel). 2024 Aug 30;14(17):2531. doi: 10.3390/ani14172531. Animals (Basel). 2024. PMID: 39272316 Free PMC article.
-
Identification of over ten thousand candidate structured RNAs in viruses and phages.Comput Struct Biotechnol J. 2023 Nov 7;21:5630-5639. doi: 10.1016/j.csbj.2023.11.010. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 38047235 Free PMC article.
-
Degenerate PCR Targeting the Major Capsid Protein Gene of HcRNAV and Related Viruses.Microbes Environ. 2022;37(5):ME21075. doi: 10.1264/jsme2.ME21075. Microbes Environ. 2022. PMID: 35400716 Free PMC article.
-
Proteome expansion in the Potyviridae evolutionary radiation.FEMS Microbiol Rev. 2022 Jul 1;46(4):fuac011. doi: 10.1093/femsre/fuac011. FEMS Microbiol Rev. 2022. PMID: 35195244 Free PMC article. Review.
-
Viral Metagenomics Reveals Widely Diverse Viral Community of Freshwater Amazonian Lake.Front Public Health. 2022 Apr 25;10:869886. doi: 10.3389/fpubh.2022.869886. eCollection 2022. Front Public Health. 2022. PMID: 35548089 Free PMC article.
References
-
- Zhang YZ, Chen YM, Wang W, Qin XC, Holmes EC. Expanding the RNA virosphere by unbiased metagenomics. Annu. Rev. Virol. 2019;6:119–139. - PubMed
-
- Lefeuvre P, et al. Evolution and ecology of plant viruses. Nat. Rev. Microbiol. 2019;17:632–644. - PubMed
-
- Obbard DJ. Expansion of the metazoan virosphere: progress, pitfalls, and prospects. Curr. Opin. Virol. 2018;31:17–23. - PubMed
-
- Brum JR, Sullivan MB. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol. 2015;13:147–159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

