Intact proviral DNA assay analysis of large cohorts of people with HIV provides a benchmark for the frequency and composition of persistent proviral DNA
- PMID: 32690683
- PMCID: PMC7414172
- DOI: 10.1073/pnas.2006816117
Intact proviral DNA assay analysis of large cohorts of people with HIV provides a benchmark for the frequency and composition of persistent proviral DNA
Abstract
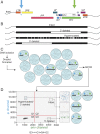
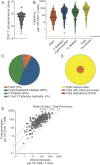
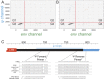
A scalable approach for quantifying intact HIV-1 proviruses is critical for basic research and clinical trials directed at HIV-1 cure. The intact proviral DNA assay (IPDA) is a novel approach to characterizing the HIV-1 reservoir, focusing on the genetic integrity of individual proviruses independent of transcriptional status. It uses multiplex digital droplet PCR to distinguish and separately quantify intact proviruses, defined by a lack of overt fatal defects such as large deletions and APOBEC3G-mediated hypermutation, from the majority of proviruses that have such defects. This distinction is important because only intact proviruses cause viral rebound on ART interruption. To evaluate IPDA performance and provide benchmark data to support its implementation, we analyzed peripheral blood samples from 400 HIV-1+ adults on ART from several diverse cohorts, representing a robust sample of treated HIV-1 infection in the United States. We provide direct quantitative evidence that defective proviruses greatly outnumber intact proviruses (by >12.5 fold). However, intact proviruses are present at substantially higher frequencies (median, 54/106 CD4+ T cells) than proviruses detected by the quantitative viral outgrowth assay, which requires induction and in vitro growth (∼1/106 CD4+ T cells). IPDA amplicon signal issues resulting from sequence polymorphisms were observed in only 6.3% of individuals and were readily apparent and easily distinguished from low proviral frequency, an advantage of the IPDA over standard PCR assays which generate false-negative results in such situations. The large IPDA dataset provided here gives the clearest quantitative picture to date of HIV-1 proviral persistence on ART.
Keywords: HIV; IPDA; cure; latency; reservoir.
Conflict of interest statement
Competing interest statement: Aspects of the IPDA are the subject of patent application PCT/US16/28822 filed by Johns Hopkins University with R.F.S. as an inventor and licensed to AccelevirDx. R.F.S. holds no equity interest in AccelevirDx. R.F.S. consults for Merck and AbbVie on HIV cure-related issues. K.D.R. and M.C. are employees of AccelevirDx. A.M. and G.M.L. are employees of and equity holders in AccelevirDx. B.J.H. is an employee of Merck.
Figures

Similar articles
-
Sequence Evaluation and Comparative Analysis of Novel Assays for Intact Proviral HIV-1 DNA.J Virol. 2021 Feb 24;95(6):e01986-20. doi: 10.1128/JVI.01986-20. Print 2021 Feb 24. J Virol. 2021. PMID: 33361426 Free PMC article.
-
Longitudinal clonal dynamics of HIV-1 latent reservoirs measured by combination quadruplex polymerase chain reaction and sequencing.Proc Natl Acad Sci U S A. 2022 Jan 25;119(4):e2117630119. doi: 10.1073/pnas.2117630119. Proc Natl Acad Sci U S A. 2022. PMID: 35042816 Free PMC article.
-
HIV-1 diversity considerations in the application of the Intact Proviral DNA Assay (IPDA).Nat Commun. 2021 Jan 8;12(1):165. doi: 10.1038/s41467-020-20442-3. Nat Commun. 2021. PMID: 33420062 Free PMC article.
-
Low Inducibility of Latent Human Immunodeficiency Virus Type 1 Proviruses as a Major Barrier to Cure.J Infect Dis. 2021 Feb 15;223(12 Suppl 2):13-21. doi: 10.1093/infdis/jiaa649. J Infect Dis. 2021. PMID: 33586775 Free PMC article. Review.
-
HIV Reservoir: How to Measure It?Curr HIV/AIDS Rep. 2023 Apr;20(2):29-41. doi: 10.1007/s11904-023-00653-1. Epub 2023 Apr 1. Curr HIV/AIDS Rep. 2023. PMID: 37004676 Review.
Cited by
-
Residual Proviral Reservoirs: A High Risk for HIV Persistence and Driving Forces for Viral Rebound after Analytical Treatment Interruption.Viruses. 2021 Feb 21;13(2):335. doi: 10.3390/v13020335. Viruses. 2021. PMID: 33670027 Free PMC article. Review.
-
Isotretinoin promotes elimination of translation-competent HIV latent reservoirs in CD4T cells.PLoS Pathog. 2024 Oct 14;20(10):e1012601. doi: 10.1371/journal.ppat.1012601. eCollection 2024 Oct. PLoS Pathog. 2024. PMID: 39401241 Free PMC article.
-
Beyond the Syndemic of Opioid Use Disorders and HIV: The Impact of Opioids on Viral Reservoirs.Viruses. 2023 Aug 9;15(8):1712. doi: 10.3390/v15081712. Viruses. 2023. PMID: 37632053 Free PMC article. Review.
-
Transient viral replication during analytical treatment interruptions in SIV infected macaques can alter the rebound-competent viral reservoir.PLoS Pathog. 2021 Jun 18;17(6):e1009686. doi: 10.1371/journal.ppat.1009686. eCollection 2021 Jun. PLoS Pathog. 2021. PMID: 34143853 Free PMC article.
-
Associations Between Multiple Measures of HIV-1 Persistence in Persons on Suppressive Antiretroviral Therapy.J Infect Dis. 2022 Jun 15;225(12):2163-2166. doi: 10.1093/infdis/jiac030. J Infect Dis. 2022. PMID: 35137129 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- UM1 AI126620/AI/NIAID NIH HHS/United States
- P30 AI094189/AI/NIAID NIH HHS/United States
- UM1 AI069481/AI/NIAID NIH HHS/United States
- P30 AI027763/AI/NIAID NIH HHS/United States
- UM1 AI106701/AI/NIAID NIH HHS/United States
- R61 DA047022/DA/NIDA NIH HHS/United States
- UM1 AI068636/AI/NIAID NIH HHS/United States
- R01 AI140789/AI/NIAID NIH HHS/United States
- U24 AI143502/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- UM1 AI126603/AI/NIAID NIH HHS/United States
- R44 AI124996/AI/NIAID NIH HHS/United States
- R43 AI142866/AI/NIAID NIH HHS/United States
- UM1 AI068634/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

