Autophagy and Tumor Database: ATdb, a novel database connecting autophagy and tumor
- PMID: 32681639
- PMCID: PMC7340339
- DOI: 10.1093/database/baaa052
Autophagy and Tumor Database: ATdb, a novel database connecting autophagy and tumor
Abstract
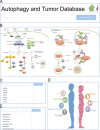
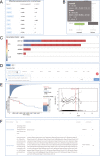
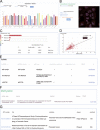
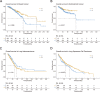
Autophagy is an essential cellular process that is closely implicated in diverse pathophysiological processes and a variety of human diseases, especially tumors. Autophagy is regarded as not only an anti-cancer process in tumorigenesis but also a pro-tumor process in progression and metastasis according to current research. It means the role of autophagy in tumor is considered to be complex, controversial and context dependent. Hence, a comprehensive database is of great significance to obtain an in-depth understanding of such complex correlations between autophagy and tumor. To achieve this objective, here we developed the Autophagy and Tumor Database (named as ATdb, http://www.bigzju.com/ATdb/#/) to compile the published information concerning autophagy and tumor research. ATdb connected 25 types of tumors with 137 genes required for autophagy-related pathways, containing 219 population filters, 2650 hazard ratio trend plots, 658 interacting microRNAs, 266 interacting long non-coding RNAs, 155 post-translational modifications, 298 DNA methylation records, 331 animal models and 70 clinical trials. ATdb could enable users to search, browse, download and carry out efficient online analysis. For instance, users can make prediction of autophagy gene regulators in a context-dependent manner and in a precise subpopulation and tumor subtypes. Also, it is feasible in ATdb to cluster tumors into distinguished groups based on the gene-related long non-coding RNAs to gain novel insights into their potential functional implications. Thus, ATdb offers a powerful online database for the autophagy community to explore the complex world of autophagy and tumor. Database URL: http://www.bigzju.com/ATdb/#/.
© The Author(s) 2020. Published by Oxford University Press.
Figures

Similar articles
-
AtDB, the Arabidopsis thaliana database, and graphical-web-display of progress by the Arabidopsis Genome Initiative.Nucleic Acids Res. 1998 Jan 1;26(1):80-4. doi: 10.1093/nar/26.1.80. Nucleic Acids Res. 1998. PMID: 9399805 Free PMC article.
-
Unified display of Arabidopsis thaliana physical maps from AtDB, the A.thaliana database.Nucleic Acids Res. 1999 Jan 1;27(1):79-84. doi: 10.1093/nar/27.1.79. Nucleic Acids Res. 1999. PMID: 9847147 Free PMC article.
-
ATDB 2.0: A database integrated toxin-ion channel interaction data.Toxicon. 2010 Sep 15;56(4):644-7. doi: 10.1016/j.toxicon.2010.05.013. Toxicon. 2010. PMID: 20677374
-
The role of epigenetics and non-coding RNAs in autophagy: A new perspective for thorough understanding.Mech Ageing Dev. 2020 Sep;190:111309. doi: 10.1016/j.mad.2020.111309. Epub 2020 Jul 4. Mech Ageing Dev. 2020. PMID: 32634442 Review.
-
Oncogenic and tumor suppressive roles of microRNAs in apoptosis and autophagy.Apoptosis. 2014 Aug;19(8):1177-89. doi: 10.1007/s10495-014-0999-7. Apoptosis. 2014. PMID: 24850099 Review.
Cited by
-
Tracing the footsteps of autophagy in computational biology.Brief Bioinform. 2021 Jul 20;22(4):bbaa286. doi: 10.1093/bib/bbaa286. Brief Bioinform. 2021. PMID: 33201177 Free PMC article.
-
A Novel Risk Model Based on Autophagy-Related LncRNAs Predicts Prognosis and Indicates Immune Infiltration Landscape of Patients With Cutaneous Melanoma.Front Genet. 2022 Apr 29;13:885391. doi: 10.3389/fgene.2022.885391. eCollection 2022. Front Genet. 2022. PMID: 35571053 Free PMC article.
-
AutophagyNet: high-resolution data source for the analysis of autophagy and its regulation.Autophagy. 2024 Jan;20(1):188-201. doi: 10.1080/15548627.2023.2247737. Epub 2023 Aug 17. Autophagy. 2024. PMID: 37589496 Free PMC article.
-
Modulating autophagy to treat diseases: A revisited review on in silico methods.J Adv Res. 2024 Apr;58:175-191. doi: 10.1016/j.jare.2023.05.002. Epub 2023 May 14. J Adv Res. 2024. PMID: 37192730 Free PMC article. Review.
-
AMTDB: A comprehensive database of autophagic modulators for anti-tumor drug discovery.Front Pharmacol. 2022 Aug 9;13:956501. doi: 10.3389/fphar.2022.956501. eCollection 2022. Front Pharmacol. 2022. PMID: 36016573 Free PMC article.
References
-
- Shen H.M. and Mizushima N. (2014) At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem. Sci., 39, 61–71. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

