Poly(A) Binding Protein Is Required for Nuclear Localization of the Ecdysteroidogenic Transcription Factor Molting Defective in the Prothoracic Gland of Drosophila melanogaster
- PMID: 32676099
- PMCID: PMC7333772
- DOI: 10.3389/fgene.2020.00636
Poly(A) Binding Protein Is Required for Nuclear Localization of the Ecdysteroidogenic Transcription Factor Molting Defective in the Prothoracic Gland of Drosophila melanogaster
Abstract
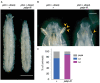
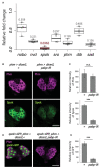
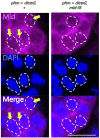
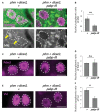
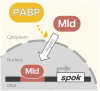
Steroid hormone signaling contributes to the development of multicellular organisms. In insects, ecdysteroids, like ecdysone and the more biologically-active derivative 20-hydroxyecdysone (20E), promote molting and metamorphosis. Ecdysone is biosynthesized in the prothoracic gland (PG), via several steps catalyzed by ecdysteroidogenic enzymes that are encoded by Halloween genes. The spatio-temporal expression pattern of ecdysteroidogenic genes is strictly controlled, resulting in a proper fluctuation of the 20E titer during insect development. However, their transcriptional regulatory mechanism is still elusive. A previous study has found that the polyadenylated tail [poly(A)] deadenylation complex, called Carbon catabolite repressor 4-Negative on TATA (CCR4-NOT) regulates the expression of spookier (spok), which encodes one of the ecdysteroidogenic enzymes in the fruit fly Drosophila melanogaster. Based on this finding, we speculated whether any other poly(A)-related protein also regulates spok expression. In this study, we reported that poly(A) binding protein (Pabp) is involved in spok expression by regulating nuclear localization of the transcription factor molting defective (Mld). When pabp was knocked down specifically in the PG by transgenic RNAi, both spok mRNA and Spok protein levels were significantly reduced. In addition, the spok promoter-driven green fluorescence protein (GFP) signal was also reduced in the pabp-RNAi PG, suggesting that Pabp is involved in the transcriptional regulation of spok. We next examined which transcription factors are responsible for Pabp-dependent transcriptional regulation. Among the transcription factors acting in the PG, we primarily focused on the zinc-finger transcription factor Mld, as Mld is essential for spok transcription. Mld was localized in the nucleus of the control PG cells, while Mld abnormally accumulated in the cytoplasm of pabp-RNAi PG cells. In contrast, pabp-RNAi did not affect the nuclear localization of other transcription factors, including ventral vein lacking (Vvl) and POU domain motif 3 (Pdm3), in PG cells. From these results, we propose that Pabp regulates subcellular localization in the PG, specifically of the transcription factor Mld, in the context of ecdysone biosynthesis.
Keywords: Halloween gene; ecdysone biosynthesis; insect development; nuclear localization; poly(A) binding protein.
Copyright © 2020 Kamiyama, Sun, Tani, Nakamura and Niwa.
Figures
Similar articles
-
Transcriptional Regulators of Ecdysteroid Biosynthetic Enzymes and Their Roles in Insect Development.Front Physiol. 2022 Feb 8;13:823418. doi: 10.3389/fphys.2022.823418. eCollection 2022. Front Physiol. 2022. PMID: 35211033 Free PMC article. Review.
-
Cooperative Control of Ecdysone Biosynthesis in Drosophila by Transcription Factors Séance, Ouija Board, and Molting Defective.Genetics. 2018 Feb;208(2):605-622. doi: 10.1534/genetics.117.300268. Epub 2017 Nov 29. Genetics. 2018. PMID: 29187506 Free PMC article.
-
Transcriptional control of steroid biosynthesis genes in the Drosophila prothoracic gland by ventral veins lacking and knirps.PLoS Genet. 2014 Jun 19;10(6):e1004343. doi: 10.1371/journal.pgen.1004343. eCollection 2014 Jun. PLoS Genet. 2014. PMID: 24945799 Free PMC article.
-
Spook and Spookier code for stage-specific components of the ecdysone biosynthetic pathway in Diptera.Dev Biol. 2006 Oct 15;298(2):555-70. doi: 10.1016/j.ydbio.2006.07.023. Epub 2006 Jul 29. Dev Biol. 2006. PMID: 16949568
-
Neural control of steroid hormone biosynthesis during development in the fruit fly Drosophila melanogaster.Genes Genet Syst. 2014;89(1):27-34. doi: 10.1266/ggs.89.27. Genes Genet Syst. 2014. PMID: 24817759 Review.
Cited by
-
Su(var)2-10- and Su(var)205-dependent upregulation of the heterochromatic gene neverland is required for developmental transition in Drosophila.Genetics. 2022 Nov 1;222(3):iyac137. doi: 10.1093/genetics/iyac137. Genetics. 2022. PMID: 36149288 Free PMC article.
-
Localization and Functional Roles of Components of the Translation Apparatus in the Eukaryotic Cell Nucleus.Cells. 2021 Nov 19;10(11):3239. doi: 10.3390/cells10113239. Cells. 2021. PMID: 34831461 Free PMC article. Review.
-
Transcriptional Regulators of Ecdysteroid Biosynthetic Enzymes and Their Roles in Insect Development.Front Physiol. 2022 Feb 8;13:823418. doi: 10.3389/fphys.2022.823418. eCollection 2022. Front Physiol. 2022. PMID: 35211033 Free PMC article. Review.
References
-
- Bellés X. (2020). Insect metamorphosis: From natural history to regulation of development and evolution. 1st Edn. Cambridge, MA, USA: Academic Press.
-
- Chávez V. M., Marqués G., Delbecque J. P., Kobayashi K., Hollingsworth M., Burr J., et al. . (2000). The Drosophila disembodied gene controls late embryonic morphogenesis and codes for a cytochrome P450 enzyme that regulates embryonic ecdysone levels. Development 127, 4115–4126. PMID: - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

