Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue
- PMID: 32675206
- PMCID: PMC7366180
- DOI: 10.1183/13993003.01123-2020
Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue
Abstract
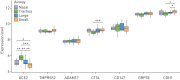
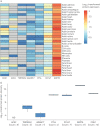
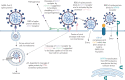
In December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged, causing the coronavirus disease 2019 (COVID-19) pandemic. SARS-CoV, the agent responsible for the 2003 SARS outbreak, utilises angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2) host molecules for viral entry. ACE2 and TMPRSS2 have recently been implicated in SARS-CoV-2 viral infection. Additional host molecules including ADAM17, cathepsin L, CD147 and GRP78 may also function as receptors for SARS-CoV-2.To determine the expression and in situ localisation of candidate SARS-CoV-2 receptors in the respiratory mucosa, we analysed gene expression datasets from airway epithelial cells of 515 healthy subjects, gene promoter activity analysis using the FANTOM5 dataset containing 120 distinct sample types, single cell RNA sequencing (scRNAseq) of 10 healthy subjects, proteomic datasets, immunoblots on multiple airway epithelial cell types, and immunohistochemistry on 98 human lung samples.We demonstrate absent to low ACE2 promoter activity in a variety of lung epithelial cell samples and low ACE2 gene expression in both microarray and scRNAseq datasets of epithelial cell populations. Consistent with gene expression, rare ACE2 protein expression was observed in the airway epithelium and alveoli of human lung, confirmed with proteomics. We present confirmatory evidence for the presence of TMPRSS2, CD147 and GRP78 protein in vitro in airway epithelial cells and confirm broad in situ protein expression of CD147 and GRP78 in the respiratory mucosa.Collectively, our data suggest the presence of a mechanism dynamically regulating ACE2 expression in human lung, perhaps in periods of SARS-CoV-2 infection, and also suggest that alternative receptors for SARS-CoV-2 exist to facilitate initial host cell infection.
Copyright ©ERS 2020.
Conflict of interest statement
Conflict of interest: J.A. Aguiar has nothing to disclose. Conflict of interest: B.J-M. Tremblay has nothing to disclose. Conflict of interest: M.J. Mansfield has nothing to disclose. Conflict of interest: O. Woody has nothing to disclose. Conflict of interest: B. Lobb has nothing to disclose. Conflict of interest: A. Banerjee has nothing to disclose. Conflict of interest: A. Chandiramohan has nothing to disclose. Conflict of interest: N. Tiessen has nothing to disclose. Conflict of interest: Q. Cao has nothing to disclose. Conflict of interest: A. Dvorkin-Gheva has nothing to disclose. Conflict of interest: S. Revill has nothing to disclose. Conflict of interest: M.S. Miller has nothing to disclose. Conflict of interest: C. Carlsten has nothing to disclose. Conflict of interest: L. Organ has nothing to disclose. Conflict of interest: C. Joseph has nothing to disclose. Conflict of interest: A. John has nothing to disclose. Conflict of interest: P. Hanson has nothing to disclose. Conflict of interest: R. Austin has a patent US7524826B2 issued to McMaster University and Hamilton Health Sciences Corp., McMaster University. Conflict of interest: B.M. McManus has nothing to disclose. Conflict of interest: G. Jenkins reports grants from AstraZeneca, Biogen, Galecto and GlaxoSmithKline; personal fees from Boehringer Ingelheim, Daewoong, Galapagos, Heptares, Promedior and Roche; grants and personal fees from Pliant; non-financial support from Redx and NuMedii; and is trustee for Action for Pulmonary Fibrosis, outside the submitted work. Conflict of interest: K. Mossman has nothing to disclose. Conflict of interest: K. Ask has nothing to disclose. Conflict of interest: A.C. Doxey has nothing to disclose. Conflict of interest: J.A. Hirota has nothing to disclose.
Figures




Similar articles
-
SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues.Cell. 2020 May 28;181(5):1016-1035.e19. doi: 10.1016/j.cell.2020.04.035. Epub 2020 Apr 27. Cell. 2020. PMID: 32413319 Free PMC article.
-
Type 2 and interferon inflammation regulate SARS-CoV-2 entry factor expression in the airway epithelium.Nat Commun. 2020 Oct 12;11(1):5139. doi: 10.1038/s41467-020-18781-2. Nat Commun. 2020. PMID: 33046696 Free PMC article.
-
Expression Pattern of the SARS-CoV-2 Entry Genes ACE2 and TMPRSS2 in the Respiratory Tract.Viruses. 2020 Oct 16;12(10):1174. doi: 10.3390/v12101174. Viruses. 2020. PMID: 33081421 Free PMC article.
-
SARS-CoV-2 pandemic and research gaps: Understanding SARS-CoV-2 interaction with the ACE2 receptor and implications for therapy.Theranostics. 2020 Jun 12;10(16):7448-7464. doi: 10.7150/thno.48076. eCollection 2020. Theranostics. 2020. PMID: 32642005 Free PMC article. Review.
-
ACE2: Evidence of role as entry receptor for SARS-CoV-2 and implications in comorbidities.Elife. 2020 Nov 9;9:e61390. doi: 10.7554/eLife.61390. Elife. 2020. PMID: 33164751 Free PMC article. Review.
Cited by
-
A basally active cGAS-STING pathway limits SARS-CoV-2 replication in a subset of ACE2 positive airway cell models.Nat Commun. 2024 Sep 27;15(1):8394. doi: 10.1038/s41467-024-52803-7. Nat Commun. 2024. PMID: 39333139 Free PMC article.
-
The Functions of SARS-CoV-2 Receptors in Diabetes-Related Severe COVID-19.Int J Mol Sci. 2024 Sep 5;25(17):9635. doi: 10.3390/ijms25179635. Int J Mol Sci. 2024. PMID: 39273582 Free PMC article. Review.
-
Bulk and Single-Cell RNA Sequencing Elucidate the Etiology of Severe COVID-19.Int J Mol Sci. 2024 Mar 14;25(6):3280. doi: 10.3390/ijms25063280. Int J Mol Sci. 2024. PMID: 38542251 Free PMC article. Review.
-
Modulations of Homeostatic ACE2, CD147, GRP78 Pathways Correlate with Vascular and Endothelial Performance Markers during Pulmonary SARS-CoV-2 Infection.Cells. 2024 Feb 29;13(5):432. doi: 10.3390/cells13050432. Cells. 2024. PMID: 38474396 Free PMC article.
-
SARS-CoV-2 mechanisms of cell tropism in various organs considering host factors.Heliyon. 2024 Feb 20;10(4):e26577. doi: 10.1016/j.heliyon.2024.e26577. eCollection 2024 Feb 29. Heliyon. 2024. PMID: 38420467 Free PMC article. Review.
References
-
- Centers for Disease Control and Prevention, National Center for Immunization and Respiratory Diseases (NCIRD) SARS Basics Fact Sheet. www.cdc.gov/sars/about/fs-sars.html Date last updated: 6 December 2017.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
