The bZIP Proteins of Oncogenic Viruses
- PMID: 32674309
- PMCID: PMC7412551
- DOI: 10.3390/v12070757
The bZIP Proteins of Oncogenic Viruses
Abstract
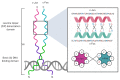
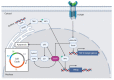
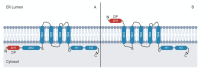
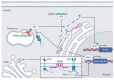
Basic leucine zipper (bZIP) transcription factors (TFs) govern diverse cellular processes and cell fate decisions. The hallmark of the leucine zipper domain is the heptad repeat, with leucine residues at every seventh position in the domain. These leucine residues enable homo- and heterodimerization between ZIP domain α-helices, generating coiled-coil structures that stabilize interactions between adjacent DNA-binding domains and target DNA substrates. Several cancer-causing viruses encode viral bZIP TFs, including human T-cell leukemia virus (HTLV), hepatitis C virus (HCV) and the herpesviruses Marek's disease virus (MDV), Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV). Here, we provide a comprehensive review of these viral bZIP TFs and their impact on viral replication, host cell responses and cell fate.
Keywords: Epstein–Barr virus (EBV); Kaposi’s sarcoma-associated herpesvirus (KSHV); Marek’s disease virus (MDV); basic leucine zipper (bZIP); hepatitis C virus (HCV); herpesvirus; human T-cell leukemia virus (HTLV).
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the writing of the manuscript.
Figures


Similar articles
-
K-bZIP of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation.J Virol. 2003 Mar;77(6):3809-15. doi: 10.1128/jvi.77.6.3809-3815.2003. J Virol. 2003. PMID: 12610155 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression.J Virol. 2003 Jan;77(2):1441-51. doi: 10.1128/jvi.77.2.1441-1451.2003. J Virol. 2003. PMID: 12502859 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus.J Virol. 1999 Mar;73(3):1909-17. doi: 10.1128/JVI.73.3.1909-1917.1999. J Virol. 1999. PMID: 9971770 Free PMC article.
-
bZIP proteins of human gammaherpesviruses.J Gen Virol. 2003 Aug;84(Pt 8):1941-1949. doi: 10.1099/vir.0.19112-0. J Gen Virol. 2003. PMID: 12867624 Review.
-
Noncoding RNAs produced by oncogenic human herpesviruses.J Cell Physiol. 2008 Aug;216(2):321-6. doi: 10.1002/jcp.21480. J Cell Physiol. 2008. PMID: 18484093 Free PMC article. Review.
Cited by
-
Identification and Functional Analysis of bZIP Genes in Cotton Response to Drought Stress.Int J Mol Sci. 2022 Nov 28;23(23):14894. doi: 10.3390/ijms232314894. Int J Mol Sci. 2022. PMID: 36499218 Free PMC article.
-
Molecular Basis of Epstein-Barr Virus Latency Establishment and Lytic Reactivation.Viruses. 2021 Nov 23;13(12):2344. doi: 10.3390/v13122344. Viruses. 2021. PMID: 34960613 Free PMC article. Review.
-
Human tumor viruses: induction of three-dimensional alterations in the host genome structure.Front Microbiol. 2023 Oct 19;14:1280210. doi: 10.3389/fmicb.2023.1280210. eCollection 2023. Front Microbiol. 2023. PMID: 37928671 Free PMC article. Review.
-
Wildebeest-Derived Malignant Catarrhal Fever: A Bovine Peripheral T Cell Lymphoma Caused by Cross-Species Transmission of Alcelaphine Gammaherpesvirus 1.Viruses. 2023 Feb 13;15(2):526. doi: 10.3390/v15020526. Viruses. 2023. PMID: 36851740 Free PMC article. Review.
-
Molecular characterization of the meq oncogene of Marek's disease virus in vaccinated Brazilian poultry farms reveals selective pressure on prevalent strains.Vet Q. 2024 Dec;44(1):1-13. doi: 10.1080/01652176.2024.2318198. Epub 2024 Mar 11. Vet Q. 2024. PMID: 38465827 Free PMC article.
References
-
- Luo Q., Viste K., Urday-Zaa J.C., Senthil Kumar G., Tsai W.-W., Talai A., Mayo K.E., Montminy M., Radhakrishnan I. Mechanism of CREB Recognition and Coactivation by the CREB-Regulated Transcriptional Coactivator CRTC2. Proc. Natl. Acad. Sci. USA. 2012;109:20865–20870. doi: 10.1073/pnas.1219028109. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

