Double-edged sword: The evolutionary consequences of the epigenetic silencing of transposable elements
- PMID: 32673310
- PMCID: PMC7365398
- DOI: 10.1371/journal.pgen.1008872
Double-edged sword: The evolutionary consequences of the epigenetic silencing of transposable elements
Abstract
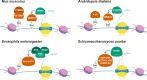
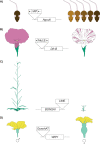
Transposable elements (TEs) are genomic parasites that selfishly replicate at the expense of host fitness. Fifty years of evolutionary studies of TEs have concentrated on the deleterious genetic effects of TEs, such as their effects on disrupting genes and regulatory sequences. However, a flurry of recent work suggests that there is another important source of TEs' harmful effects-epigenetic silencing. Host genomes typically silence TEs by the deposition of repressive epigenetic marks. While this silencing reduces the selfish replication of TEs and should benefit hosts, a picture is emerging that the epigenetic silencing of TEs triggers inadvertent spreading of repressive marks to otherwise expressed neighboring genes, ultimately jeopardizing host fitness. In this Review, we provide a long-overdue overview of the recent genome-wide evidence for the presence and prevalence of TEs' epigenetic effects, highlighting both the similarities and differences across mammals, insects, and plants. We lay out the current understanding of the functional and fitness consequences of TEs' epigenetic effects, and propose possible influences of such effects on the evolution of both hosts and TEs themselves. These unique evolutionary consequences indicate that TEs' epigenetic effect is not only a crucial component of TE biology but could also be a significant contributor to genome function and evolution.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Species-specific chromatin landscape determines how transposable elements shape genome evolution.Elife. 2022 Aug 23;11:e81567. doi: 10.7554/eLife.81567. Elife. 2022. PMID: 35997258 Free PMC article.
-
Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression.Genome Res. 2009 Aug;19(8):1419-28. doi: 10.1101/gr.091678.109. Epub 2009 May 28. Genome Res. 2009. PMID: 19478138 Free PMC article.
-
Epigenetic regulation of intragenic transposable elements: a two-edged sword.J Biochem. 2018 Nov 1;164(5):323-328. doi: 10.1093/jb/mvy060. J Biochem. 2018. PMID: 30010918 Review.
-
Blessing or curse: how the epigenetic resolution of host-transposable element conflicts shapes their evolutionary dynamics.Proc Biol Sci. 2024 Apr 10;291(2020):20232775. doi: 10.1098/rspb.2023.2775. Epub 2024 Apr 10. Proc Biol Sci. 2024. PMID: 38593848 Free PMC article.
-
Epigenetic regulation and functional exaptation of transposable elements in higher plants.Curr Opin Plant Biol. 2014 Oct;21:83-88. doi: 10.1016/j.pbi.2014.07.001. Epub 2014 Jul 25. Curr Opin Plant Biol. 2014. PMID: 25061895 Review.
Cited by
-
Taming, Domestication and Exaptation: Trajectories of Transposable Elements in Genomes.Cells. 2021 Dec 20;10(12):3590. doi: 10.3390/cells10123590. Cells. 2021. PMID: 34944100 Free PMC article. Review.
-
The Tetragnatha kauaiensis Genome Sheds Light on the Origins of Genomic Novelty in Spiders.Genome Biol Evol. 2021 Dec 1;13(12):evab262. doi: 10.1093/gbe/evab262. Genome Biol Evol. 2021. PMID: 34849853 Free PMC article.
-
CHH Methylation Islands: A Nonconserved Feature of Grass Genomes That Is Positively Associated with Transposable Elements but Negatively Associated with Gene-Body Methylation.Genome Biol Evol. 2021 Aug 3;13(8):evab144. doi: 10.1093/gbe/evab144. Genome Biol Evol. 2021. PMID: 34146109 Free PMC article.
-
Satellite DNA-Mediated Gene Expression Regulation: Physiological and Evolutionary Implication.Prog Mol Subcell Biol. 2021;60:145-167. doi: 10.1007/978-3-030-74889-0_6. Prog Mol Subcell Biol. 2021. PMID: 34386875
-
Characterization of genomic regions escaping epigenetic reprogramming in sheep.Environ Epigenet. 2023 Dec 20;10(1):dvad010. doi: 10.1093/eep/dvad010. eCollection 2024. Environ Epigenet. 2023. PMID: 38496251 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

