A Rare Mutation in an Infant-Derived HIV-1 Envelope Glycoprotein Alters Interprotomer Stability and Susceptibility to Broadly Neutralizing Antibodies Targeting the Trimer Apex
- PMID: 32669335
- PMCID: PMC7495384
- DOI: 10.1128/JVI.00814-20
A Rare Mutation in an Infant-Derived HIV-1 Envelope Glycoprotein Alters Interprotomer Stability and Susceptibility to Broadly Neutralizing Antibodies Targeting the Trimer Apex
Abstract
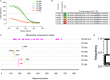
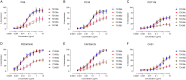
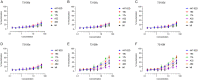
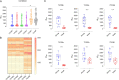
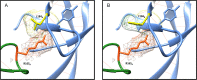
The envelope glycoprotein (Env) of human immunodeficiency virus type 1 (HIV-1) is the sole target of broadly neutralizing antibodies (bnAbs). Several mechanisms, such as the acquisition of mutations, variability of the loop length, and alterations in the glycan pattern, are employed by the virus to shield neutralizing epitopes on Env to sustain survival and infectivity within the host. The identification of mutations that lead to viral evasion of the host immune response is essential for the optimization and engineering of Env-based trimeric immunogens. Here, we report a rare leucine-to-phenylalanine escape mutation (L184F) at the base of hypervariable loop 2 (population frequency of 0.0045%) in a 9-month-old perinatally HIV-1-infected infant broad neutralizer. The L184F mutation altered the trimer conformation by modulating intramolecular interactions stabilizing the trimer apex and led to viral escape from autologous plasma bnAbs and known N160 glycan-targeted bnAbs. The L184F amino acid change led to the acquisition of a relatively open trimeric conformation, often associated with tier 1 HIV-1 isolates and increased susceptibility to neutralization by polyclonal plasma antibodies of weak neutralizers. While there was no impact of the L184F mutation on free virus transmission, a reduction in cell-to-cell transmission was observed. In conclusion, we report a naturally selected viral mutation, L184F, that influenced a change in the conformation of the Env trimer apex as a mechanism of escape from contemporaneous plasma V2 apex-targeted nAbs. Further studies should be undertaken to define viral mutations acquired during natural infection, to escape selection pressure exerted by bnAbs, to inform vaccine design and bnAb-based therapeutic strategies.IMPORTANCE The design of HIV-1 envelope-based immunogens capable of eliciting broadly neutralizing antibodies (bnAbs) is currently under active research. Some of the most potent bnAbs target the quaternary epitope at the V2 apex of the HIV-1 Env trimer. By studying naturally circulating viruses from a perinatally HIV-1-infected infant with plasma neutralizing antibodies targeted to the V2 apex, we identified a rare leucine-to-phenylalanine substitution, in two out of six functional viral clones, that destabilized the trimer apex. This single-amino-acid alteration impaired the interprotomeric interactions that stabilize the trimer apex, resulting in an open trimer conformation and escape from broadly neutralizing autologous plasma antibodies and known V2 apex-directed bnAbs, thereby favoring viral evasion of the early bnAb response of the infected host. Defining the mechanisms by which naturally occurring viral mutations influence the sensitivity of HIV-1 to bnAbs will provide information for the development of vaccines and bnAbs as anti-HIV-1 reagents.
Keywords: HIV-1; V2 apex bnAbs; infants; interprotomer interactions; rare mutation.
Copyright © 2020 American Society for Microbiology.
Figures



Similar articles
-
Functional Stability of HIV-1 Envelope Trimer Affects Accessibility to Broadly Neutralizing Antibodies at Its Apex.J Virol. 2017 Nov 30;91(24):e01216-17. doi: 10.1128/JVI.01216-17. Print 2017 Dec 15. J Virol. 2017. PMID: 28978711 Free PMC article.
-
Structural Constraints at the Trimer Apex Stabilize the HIV-1 Envelope in a Closed, Antibody-Protected Conformation.mBio. 2018 Dec 11;9(6):e00955-18. doi: 10.1128/mBio.00955-18. mBio. 2018. PMID: 30538178 Free PMC article.
-
Conformational Epitope-Specific Broadly Neutralizing Plasma Antibodies Obtained from an HIV-1 Clade C-Infected Elite Neutralizer Mediate Autologous Virus Escape through Mutations in the V1 Loop.J Virol. 2016 Jan 13;90(7):3446-57. doi: 10.1128/JVI.03090-15. J Virol. 2016. PMID: 26763999 Free PMC article.
-
Stabilizing HIV-1 envelope glycoprotein trimers to induce neutralizing antibodies.Retrovirology. 2018 Sep 12;15(1):63. doi: 10.1186/s12977-018-0445-y. Retrovirology. 2018. PMID: 30208933 Free PMC article. Review.
-
Strategies for induction of HIV-1 envelope-reactive broadly neutralizing antibodies.J Int AIDS Soc. 2021 Nov;24 Suppl 7(Suppl 7):e25831. doi: 10.1002/jia2.25831. J Int AIDS Soc. 2021. PMID: 34806332 Free PMC article. Review.
Cited by
-
Recognition determinants of improved HIV-1 neutralization by a heavy chain matured pediatric antibody.iScience. 2023 Aug 9;26(9):107579. doi: 10.1016/j.isci.2023.107579. eCollection 2023 Sep 15. iScience. 2023. PMID: 37649696 Free PMC article.
-
Global Increases in Human Immunodeficiency Virus Neutralization Sensitivity Due to Alterations in the Membrane-Proximal External Region of the Envelope Glycoprotein Can Be Minimized by Distant State 1-Stabilizing Changes.J Virol. 2022 Apr 13;96(7):e0187821. doi: 10.1128/jvi.01878-21. Epub 2022 Mar 15. J Virol. 2022. PMID: 35289647 Free PMC article.
-
B cell repertoire sequencing of HIV-1 pediatric elite-neutralizers identifies multiple broadly neutralizing antibody clonotypes.Front Immunol. 2024 Feb 16;15:1272493. doi: 10.3389/fimmu.2024.1272493. eCollection 2024. Front Immunol. 2024. PMID: 38433846 Free PMC article.
-
Regulation of epitope exposure in the gp41 membrane-proximal external region through interactions at the apex of HIV-1 Env.PLoS Pathog. 2022 May 18;18(5):e1010531. doi: 10.1371/journal.ppat.1010531. eCollection 2022 May. PLoS Pathog. 2022. PMID: 35584191 Free PMC article.
-
An Overview of Human Anti-HIV-1 Neutralizing Antibodies against Diverse Epitopes of HIV-1.ACS Omega. 2023 Feb 15;8(8):7252-7261. doi: 10.1021/acsomega.2c07933. eCollection 2023 Feb 28. ACS Omega. 2023. PMID: 36873012 Free PMC article. Review.
References
-
- Steichen JM, Lin Y-C, Havenar-Daughton C, Pecetta S, Ozorowski G, Willis JR, Toy L, Sok D, Liguori A, Kratochvil S, Torres JL, Kalyuzhniy O, Melzi E, Kulp DW, Raemisch S, Hu X, Bernard SM, Georgeson E, Phelps N, Adachi Y, Kubitz M, Landais E, Umotoy J, Robinson A, Briney B, Wilson IA, Burton DR, Ward AB, Crotty S, Batista FD, Schief WR. 2019. A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses. Science 366:eaax4380. doi:10.1126/science.aax4380. - DOI - PMC - PubMed
-
- Saunders KO, Wiehe K, Tian M, Acharya P, Bradley T, Alam SM, Go EP, Scearce R, Sutherland L, Henderson R, Hsu AL, Borgnia MJ, Chen H, Lu X, Wu NR, Watts B, Jiang C, Easterhoff D, Cheng H-L, McGovern K, Waddicor P, Chapdelaine-Williams A, Eaton A, Zhang J, Rountree W, Verkoczy L, Tomai M, Lewis MG, Desaire HR, Edwards RJ, Cain DW, Bonsignori M, Montefiori D, Alt FW, Haynes BF. 2019. Targeted selection of HIV-specific antibody mutations by engineering B cell maturation. Science 366:eaay7199. doi:10.1126/science.aay7199. - DOI - PMC - PubMed
-
- Ghulam-Smith M, Olson A, White LF, Chasela CS, Ellington SR, Kourtis AP, Jamieson DJ, Tegha G, van der Horst CM, Sagar M. 2017. Maternal but not infant anti-HIV-1 neutralizing antibody response associates with enhanced transmission and infant morbidity. mBio 8:e01373-17. doi:10.1128/mBio.01373-17. - DOI - PMC - PubMed
-
- Kumar A, Smith CEP, Giorgi EE, Eudailey J, Martinez DR, Yusim K, Douglas AO, Stamper L, McGuire E, LaBranche CC, Montefiori DC, Fouda GG, Gao F, Permar SR. 2018. Infant transmitted/founder HIV-1 viruses from peripartum transmission are neutralization resistant to paired maternal plasma. PLoS Pathog 14:e1006944. doi:10.1371/journal.ppat.1006944. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

