The pregnant myometrium is epigenetically activated at contractility-driving gene loci prior to the onset of labor in mice
- PMID: 32667910
- PMCID: PMC7384763
- DOI: 10.1371/journal.pbio.3000710
The pregnant myometrium is epigenetically activated at contractility-driving gene loci prior to the onset of labor in mice
Abstract
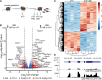
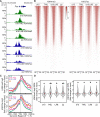
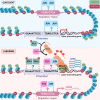
During gestation, uterine smooth muscle cells transition from a state of quiescence to one of contractility, but the molecular mechanisms underlying this transition at a genomic level are not well-known. To better understand these events, we evaluated the epigenetic landscape of the mouse myometrium during the pregnant, laboring, and postpartum stages. We generated gestational time point-specific enrichment profiles for histone H3 acetylation on lysine residue 27 (H3K27ac), histone H3 trimethylation of lysine residue 4 (H3K4me3), and RNA polymerase II (RNAPII) occupancy by chromatin immunoprecipitation with massively parallel sequencing (ChIP-seq), as well as gene expression profiles by total RNA-sequencing (RNA-seq). Our findings reveal that 533 genes, including known contractility-driving genes (Gap junction alpha 1 [Gja1], FBJ osteosarcoma oncogene [Fos], Fos-like antigen 2 [Fosl2], Oxytocin receptor [Oxtr], and Prostaglandin G/H synthase 2 (Ptgs2), for example), are up-regulated at day 19 during active labor because of an increase in transcription at gene bodies. Labor-associated promoters and putative intergenic enhancers, however, are epigenetically activated as early as day 15, by which point the majority of genome-wide H3K27ac or H3K4me3 peaks present in term laboring tissue is already established. Despite this early exhibited histone signature, increased noncoding enhancer RNA (eRNA) production at putative intergenic enhancers and recruitment of RNAPII to the gene bodies of labor-associated loci were detected only during labor. Our findings indicate that epigenetic activation of the myometrial genome precedes active labor by at least 4 days in the mouse model, suggesting that the myometrium is poised for rapid activation of contraction-associated genes in order to exit the state of quiescence.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Epigenetics of human myometrium: DNA methylation of genes encoding contraction-associated proteins in term and preterm labor.Biol Reprod. 2014 May 8;90(5):98. doi: 10.1095/biolreprod.113.113209. Print 2014 May. Biol Reprod. 2014. PMID: 24571989 Free PMC article.
-
Up-regulation of the progesterone receptor (PR)-C isoform in laboring myometrium by activation of nuclear factor-kappaB may contribute to the onset of labor through inhibition of PR function.Mol Endocrinol. 2006 Apr;20(4):764-75. doi: 10.1210/me.2005-0242. Epub 2005 Dec 8. Mol Endocrinol. 2006. PMID: 16339279
-
Term myometrium is characterized by increased activating epigenetic modifications at the progesterone receptor-A promoter.Mol Hum Reprod. 2012 Aug;18(8):401-9. doi: 10.1093/molehr/gas012. Epub 2012 Feb 27. Mol Hum Reprod. 2012. PMID: 22369759
-
Biochemistry of myometrial contractility.Baillieres Clin Obstet Gynaecol. 1992 Dec;6(4):755-69. doi: 10.1016/s0950-3552(05)80187-7. Baillieres Clin Obstet Gynaecol. 1992. PMID: 1335853 Review.
-
Evaluating Enhancer Function and Transcription.Annu Rev Biochem. 2020 Jun 20;89:213-234. doi: 10.1146/annurev-biochem-011420-095916. Epub 2020 Mar 20. Annu Rev Biochem. 2020. PMID: 32197056 Review.
Cited by
-
MYB and ELF3 differentially modulate labor-inducing gene expression in myometrial cells.PLoS One. 2023 Jan 3;18(1):e0271081. doi: 10.1371/journal.pone.0271081. eCollection 2023. PLoS One. 2023. PMID: 36595497 Free PMC article.
-
Stromal cells-specific retinoic acid determines parturition timing at single-cell and spatial-temporal resolution.iScience. 2023 Sep 1;26(10):107796. doi: 10.1016/j.isci.2023.107796. eCollection 2023 Oct 20. iScience. 2023. PMID: 37720083 Free PMC article.
-
Decline in corepressor CNOT1 in the pregnant myometrium near term impairs progesterone receptor function and increases contractile gene expression.J Biol Chem. 2024 Jul;300(7):107484. doi: 10.1016/j.jbc.2024.107484. Epub 2024 Jun 18. J Biol Chem. 2024. PMID: 38897566 Free PMC article.
-
Prostaglandin F2α requires activation of calcium-dependent signalling to trigger inflammation in human myometrium.Front Endocrinol (Lausanne). 2023 Jul 19;14:1150125. doi: 10.3389/fendo.2023.1150125. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37547305 Free PMC article.
-
Next generation strategies for preventing preterm birth.Adv Drug Deliv Rev. 2021 Jul;174:190-209. doi: 10.1016/j.addr.2021.04.021. Epub 2021 Apr 23. Adv Drug Deliv Rev. 2021. PMID: 33895215 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

