A trafficome-wide RNAi screen reveals deployment of early and late secretory host proteins and the entire late endo-/lysosomal vesicle fusion machinery by intracellular Salmonella
- PMID: 32658937
- PMCID: PMC7377517
- DOI: 10.1371/journal.ppat.1008220
A trafficome-wide RNAi screen reveals deployment of early and late secretory host proteins and the entire late endo-/lysosomal vesicle fusion machinery by intracellular Salmonella
Abstract
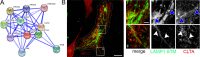
The intracellular lifestyle of Salmonella enterica is characterized by the formation of a replication-permissive membrane-bound niche, the Salmonella-containing vacuole (SCV). As a further consequence of the massive remodeling of the host cell endosomal system, intracellular Salmonella establish a unique network of various Salmonella-induced tubules (SIT). The bacterial repertoire of effector proteins required for the establishment for one type of these SIT, the Salmonella-induced filaments (SIF), is rather well-defined. However, the corresponding host cell proteins are still poorly understood. To identify host factors required for the formation of SIF, we performed a sub-genomic RNAi screen. The analyses comprised high-resolution live cell imaging to score effects on SIF induction, dynamics and morphology. The hits of our functional RNAi screen comprise: i) The late endo-/lysosomal SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex, consisting of STX7, STX8, VTI1B, and VAMP7 or VAMP8, which is, in conjunction with RAB7 and the homotypic fusion and protein sorting (HOPS) tethering complex, a complete vesicle fusion machinery. ii) Novel interactions with the early secretory GTPases RAB1A and RAB1B, providing a potential link to coat protein complex I (COPI) vesicles and reinforcing recently identified ties to the endoplasmic reticulum. iii) New connections to the late secretory pathway and/or the recycling endosome via the GTPases RAB3A, RAB8A, and RAB8B and the SNAREs VAMP2, VAMP3, and VAMP4. iv) An unprecedented involvement of clathrin-coated structures. The resulting set of hits allowed us to characterize completely new host factor interactions, and to strengthen observations from several previous studies.
Conflict of interest statement
No authors have competing interests
Figures


Similar articles
-
Salmonella exploits the host endolysosomal tethering factor HOPS complex to promote its intravacuolar replication.PLoS Pathog. 2017 Oct 30;13(10):e1006700. doi: 10.1371/journal.ppat.1006700. eCollection 2017 Oct. PLoS Pathog. 2017. PMID: 29084291 Free PMC article.
-
Live cell imaging reveals novel functions of Salmonella enterica SPI2-T3SS effector proteins in remodeling of the host cell endosomal system.PLoS One. 2014 Dec 18;9(12):e115423. doi: 10.1371/journal.pone.0115423. eCollection 2014. PLoS One. 2014. PMID: 25522146 Free PMC article.
-
The delivery of endocytosed cargo to lysosomes.Biochem Soc Trans. 2009 Oct;37(Pt 5):1019-21. doi: 10.1042/BST0371019. Biochem Soc Trans. 2009. PMID: 19754443 Review.
-
Multiple Salmonella-pathogenicity island 2 effectors are required to facilitate bacterial establishment of its intracellular niche and virulence.PLoS One. 2020 Jun 25;15(6):e0235020. doi: 10.1371/journal.pone.0235020. eCollection 2020. PLoS One. 2020. PMID: 32584855 Free PMC article.
-
Take the tube: remodelling of the endosomal system by intracellular Salmonella enterica.Cell Microbiol. 2015 May;17(5):639-47. doi: 10.1111/cmi.12441. Epub 2015 Apr 10. Cell Microbiol. 2015. PMID: 25802001 Review.
Cited by
-
Salmonella Exhibit Altered Cellular Localization in the Presence of HLA-B27 and Codistribute with Endo-Reticular Membrane.J Immunol Res. 2022 Sep 16;2022:9493019. doi: 10.1155/2022/9493019. eCollection 2022. J Immunol Res. 2022. PMID: 36157878 Free PMC article.
-
The multifaceted interactions between pathogens and host ESCRT machinery.PLoS Pathog. 2023 May 4;19(5):e1011344. doi: 10.1371/journal.ppat.1011344. eCollection 2023 May. PLoS Pathog. 2023. PMID: 37141275 Free PMC article. Review.
-
SAC1 regulates autophagosomal phosphatidylinositol-4-phosphate for xenophagy-directed bacterial clearance.Cell Rep. 2021 Jul 27;36(4):109434. doi: 10.1016/j.celrep.2021.109434. Cell Rep. 2021. PMID: 34320354 Free PMC article.
-
A host E3 ubiquitin ligase regulates Salmonella virulence by targeting an SPI-2 effector involved in SIF biogenesis.mLife. 2023 Jun 13;2(2):141-158. doi: 10.1002/mlf2.12063. eCollection 2023 Jun. mLife. 2023. PMID: 38817622 Free PMC article.
-
Predicting host dependency factors of pathogens in Drosophila melanogaster using machine learning.Comput Struct Biotechnol J. 2021 Aug 9;19:4581-4592. doi: 10.1016/j.csbj.2021.08.010. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34471501 Free PMC article.
References
-
- Ramos-Morales F. Impact of Salmonella enterica Type III Secretion System Effectors on the Eukaryotic Host Cell. ISRN Cell Biology. 2012;2012 10.5402/2012/787934 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

