Identification and characterization of adipose surface epitopes
- PMID: 32648930
- PMCID: PMC7360119
- DOI: 10.1042/BCJ20190462
Identification and characterization of adipose surface epitopes
Abstract
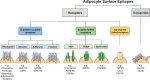
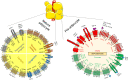
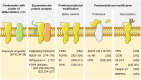
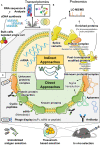
Adipose tissue is a central regulator of metabolism and an important pharmacological target to treat the metabolic consequences of obesity, such as insulin resistance and dyslipidemia. Among the various cellular compartments, the adipocyte cell surface is especially appealing as a drug target as it contains various proteins that when activated or inhibited promote adipocyte health, change its endocrine function and eventually maintain or restore whole-body insulin sensitivity. In addition, cell surface proteins are readily accessible by various drug classes. However, targeting individual cell surface proteins in adipocytes has been difficult due to important functions of these proteins outside adipose tissue, raising various safety concerns. Thus, one of the biggest challenges is the lack of adipose selective surface proteins and/or targeting reagents. Here, we discuss several receptor families with an important function in adipogenesis and mature adipocytes to highlight the complexity at the cell surface and illustrate the problems with identifying adipose selective proteins. We then discuss that, while no unique adipocyte surface protein might exist, how splicing, posttranslational modifications as well as protein/protein interactions can create enormous diversity at the cell surface that vastly expands the space of potentially unique epitopes and how these selective epitopes can be identified and targeted.
Keywords: adipogenesis; brown adipose tissue; insulin resistance; obesity; surface markers; white adipose tissue.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures
Similar articles
-
Adipocyte-specific deletion of mTOR inhibits adipose tissue development and causes insulin resistance in mice.Diabetologia. 2016 Sep;59(9):1995-2004. doi: 10.1007/s00125-016-4006-4. Epub 2016 Jun 13. Diabetologia. 2016. PMID: 27294611 Free PMC article.
-
Lack of Gαi2 proteins in adipocytes attenuates diet-induced obesity.Mol Metab. 2020 Oct;40:101029. doi: 10.1016/j.molmet.2020.101029. Epub 2020 May 30. Mol Metab. 2020. PMID: 32480042 Free PMC article.
-
Adipocyte lineages: tracing back the origins of fat.Biochim Biophys Acta. 2014 Mar;1842(3):340-51. doi: 10.1016/j.bbadis.2013.05.027. Epub 2013 Jun 4. Biochim Biophys Acta. 2014. PMID: 23747579 Free PMC article. Review.
-
Adipocyte-specific Hypoxia-inducible gene 2 promotes fat deposition and diet-induced insulin resistance.Mol Metab. 2016 Sep 28;5(12):1149-1161. doi: 10.1016/j.molmet.2016.09.009. eCollection 2016 Dec. Mol Metab. 2016. PMID: 27900258 Free PMC article.
-
Adipose tissue plasticity from WAT to BAT and in between.Biochim Biophys Acta. 2014 Mar;1842(3):358-69. doi: 10.1016/j.bbadis.2013.05.011. Epub 2013 May 17. Biochim Biophys Acta. 2014. PMID: 23688783 Free PMC article. Review.
Cited by
-
Identification of Novel Ligands for Targeted Antifibrotic Therapy of Chronic Pancreatitis.Int J Nanomedicine. 2021 Aug 14;16:5495-5512. doi: 10.2147/IJN.S318331. eCollection 2021. Int J Nanomedicine. 2021. PMID: 34429596 Free PMC article.
-
Identification and characterization of distinct brown adipocyte subtypes in C57BL/6J mice.Life Sci Alliance. 2020 Nov 30;4(1):e202000924. doi: 10.26508/lsa.202000924. Print 2021 Jan. Life Sci Alliance. 2020. PMID: 33257475 Free PMC article.
-
Immune Cell Regulation of White Adipose Progenitor Cell Fate.Front Endocrinol (Lausanne). 2022 Mar 29;13:859044. doi: 10.3389/fendo.2022.859044. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35422761 Free PMC article. Review.
-
Chemically Defined Xeno- and Serum-Free Cell Culture Medium to Grow Human Adipose Stem Cells.Cells. 2021 Feb 22;10(2):466. doi: 10.3390/cells10020466. Cells. 2021. PMID: 33671568 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

