Analysis of CRISPR/Cas9 Guide RNA Efficiency and Specificity Against Genetically Diverse HIV-1 Isolates
- PMID: 32640832
- PMCID: PMC7549012
- DOI: 10.1089/AID.2020.0055
Analysis of CRISPR/Cas9 Guide RNA Efficiency and Specificity Against Genetically Diverse HIV-1 Isolates
Abstract
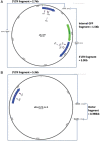
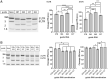
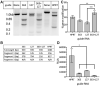
Gene editing approaches using CRISPR/Cas9 are being developed as a means for targeting the integrated HIV-1 provirus. Enthusiasm for the use of gene editing as an anti-HIV-1 therapeutic has been tempered by concerns about the specificity and efficacy of this approach. Guide RNAs (gRNAs) that target conserved sequences across a wide range of genetically diverse HIV-1 isolates will have greater clinical utility. However, on-target efficacy should be considered in the context of off-target cleavage events as these may comprise an essential safety parameter for CRISPR-based therapeutics. We analyzed a panel of Streptococcus pyogenes Cas9 (SpCas9) gRNAs directed to the 5' and 3' long terminal repeat (LTR) regions of HIV-1. We used in vitro cleavage assays with genetically diverse HIV-1 LTR sequences to determine gRNA activity across HIV-1 clades. Lipid-based transfection of gRNA/Cas9 ribonucleoproteins was used to assess targeting of the integrated HIV-1 proviral sequence in cells (in vivo). For both the in vitro and in vivo experiments, we observed increased efficiency of sequence disruption through the simultaneous use of two distinct gRNAs. Next, CIRCLE-Seq was utilized to identify off-target cleavage events using genomic DNA from cells with integrated HIV-1 proviral DNA. We identified a gRNA targeting the U3 region of the LTR (termed SpCas9-127HBX2) with broad cleavage efficiency against sequences from genetically diverse HIV-1 strains. Based on these results, we propose a workflow for identification and development of anti-HIV CRISPR therapeutics.
Keywords: CRIPSR; Cas9; HIV-1; HIV-1 LTR; off-target analysis.
Conflict of interest statement
No competing financial interests exist.
Figures



Similar articles
-
Gene Therapy with CRISPR/Cas9 Coming to Age for HIV Cure.AIDS Rev. 2017 Oct-Dec;19(3):167-172. AIDS Rev. 2017. PMID: 29019352
-
Broad-Spectrum and Personalized Guide RNAs for CRISPR/Cas9 HIV-1 Therapeutics.AIDS Res Hum Retroviruses. 2018 Nov;34(11):950-960. doi: 10.1089/AID.2017.0274. Epub 2018 Aug 27. AIDS Res Hum Retroviruses. 2018. PMID: 29968495 Free PMC article.
-
Novel gRNA design pipeline to develop broad-spectrum CRISPR/Cas9 gRNAs for safe targeting of the HIV-1 quasispecies in patients.Sci Rep. 2019 Nov 19;9(1):17088. doi: 10.1038/s41598-019-52353-9. Sci Rep. 2019. PMID: 31745112 Free PMC article.
-
CRISPR-CAS Replacing Antiviral Drugs against HIV: An Update.Crit Rev Eukaryot Gene Expr. 2020;30(1):77-83. doi: 10.1615/CritRevEukaryotGeneExpr.2020028233. Crit Rev Eukaryot Gene Expr. 2020. PMID: 32421986 Review.
-
Genome Editing with CRISPR-Cas9: Can It Get Any Better?J Genet Genomics. 2016 May 20;43(5):239-50. doi: 10.1016/j.jgg.2016.04.008. Epub 2016 Apr 24. J Genet Genomics. 2016. PMID: 27210042 Free PMC article. Review.
Cited by
-
Computational analysis of cas proteins unlocks new potential in HIV-1 targeted gene therapy.Front Genome Ed. 2024 Jan 4;5:1248982. doi: 10.3389/fgeed.2023.1248982. eCollection 2023. Front Genome Ed. 2024. PMID: 38239625 Free PMC article.
-
CRISPR/Cas9 Ablation of Integrated HIV-1 Accumulates Proviral DNA Circles with Reformed Long Terminal Repeats.J Virol. 2021 Nov 9;95(23):e0135821. doi: 10.1128/JVI.01358-21. Epub 2021 Sep 22. J Virol. 2021. PMID: 34549986 Free PMC article.
-
Updates on CRISPR-based gene editing in HIV-1/AIDS therapy.Virol Sin. 2022 Feb;37(1):1-10. doi: 10.1016/j.virs.2022.01.017. Epub 2022 Jan 19. Virol Sin. 2022. PMID: 35234622 Free PMC article. Review.
-
CRISPR-Cas9 Mediated Exonic Disruption for HIV-1 Elimination.EBioMedicine. 2021 Nov;73:103678. doi: 10.1016/j.ebiom.2021.103678. Epub 2021 Nov 10. EBioMedicine. 2021. PMID: 34774454 Free PMC article.
-
Pathways Toward a Functional HIV-1 Cure: Balancing Promise and Perils of CRISPR Therapy.Methods Mol Biol. 2022;2407:429-445. doi: 10.1007/978-1-0716-1871-4_27. Methods Mol Biol. 2022. PMID: 34985679 Free PMC article. Review.
References
-
- de Buhr H, Lebbink RJ: Harnessing CRISPR to combat human viral infections. Curr Opin Immunol 2018;54:123–129 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

