A Critical Review of Bottom-Up Proteomics: The Good, the Bad, and the Future of this Field
- PMID: 32640657
- PMCID: PMC7564415
- DOI: 10.3390/proteomes8030014
A Critical Review of Bottom-Up Proteomics: The Good, the Bad, and the Future of this Field
Abstract
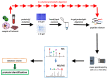
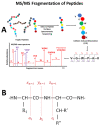
Proteomics is the field of study that includes the analysis of proteins, from either a basic science prospective or a clinical one. Proteins can be investigated for their abundance, variety of proteoforms due to post-translational modifications (PTMs), and their stable or transient protein-protein interactions. This can be especially beneficial in the clinical setting when studying proteins involved in different diseases and conditions. Here, we aim to describe a bottom-up proteomics workflow from sample preparation to data analysis, including all of its benefits and pitfalls. We also describe potential improvements in this type of proteomics workflow for the future.
Keywords: mass spectrometry; protein characterization; protein identification; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Top-Down Proteomics Applied to Human Cerebrospinal Fluid.Methods Mol Biol. 2019;2044:193-219. doi: 10.1007/978-1-4939-9706-0_12. Methods Mol Biol. 2019. PMID: 31432414
-
Top Down Proteomics Reveals Mature Proteoforms Expressed in Subcellular Fractions of the Echinococcus granulosus Preadult Stage.J Proteome Res. 2015 Nov 6;14(11):4805-14. doi: 10.1021/acs.jproteome.5b00642. Epub 2015 Oct 28. J Proteome Res. 2015. PMID: 26465659 Free PMC article.
-
Characterization of Proteoforms with Unknown Post-translational Modifications Using the MIScore.J Proteome Res. 2016 Aug 5;15(8):2422-32. doi: 10.1021/acs.jproteome.5b01098. Epub 2016 Jul 1. J Proteome Res. 2016. PMID: 27291504 Free PMC article.
-
Profiling proteoforms: promising follow-up of proteomics for biomarker discovery.Expert Rev Proteomics. 2014 Feb;11(1):121-9. doi: 10.1586/14789450.2014.878652. Expert Rev Proteomics. 2014. PMID: 24437377 Review.
-
Comprehensive Overview of Bottom-Up Proteomics Using Mass Spectrometry.ACS Meas Sci Au. 2024 Jun 4;4(4):338-417. doi: 10.1021/acsmeasuresciau.3c00068. eCollection 2024 Aug 21. ACS Meas Sci Au. 2024. PMID: 39193565 Free PMC article. Review.
Cited by
-
Clinical Implications of a Proteomics-Based Approach for Cardiomyopathy and Heart Failure.Korean Circ J. 2024 Aug;54(8):482-484. doi: 10.4070/kcj.2024.0194. Korean Circ J. 2024. PMID: 39109596 Free PMC article. No abstract available.
-
PANAMA-enabled high-sensitivity dual nanoflow LC-MS metabolomics and proteomics analysis.Cell Rep Methods. 2024 Jul 15;4(7):100803. doi: 10.1016/j.crmeth.2024.100803. Epub 2024 Jul 2. Cell Rep Methods. 2024. PMID: 38959888 Free PMC article.
-
Proteoforms expand the world of microproteins and short open reading frame-encoded peptides.iScience. 2023 Jan 27;26(2):106069. doi: 10.1016/j.isci.2023.106069. eCollection 2023 Feb 17. iScience. 2023. PMID: 36818287 Free PMC article. Review.
-
A Deep Convolutional Neural Network for Prediction of Peptide Collision Cross Sections in Ion Mobility Spectrometry.Biomolecules. 2021 Dec 19;11(12):1904. doi: 10.3390/biom11121904. Biomolecules. 2021. PMID: 34944547 Free PMC article.
-
Proteomic Advances in Cereal and Vegetable Crops.Molecules. 2021 Aug 14;26(16):4924. doi: 10.3390/molecules26164924. Molecules. 2021. PMID: 34443513 Free PMC article. Review.
References
-
- Schwenk J.M., Omenn G.S., Sun Z., Campbell D.S., Baker M.S., Overall C.M., Aebersold R., Moritz R.L., Deutsch E.W. The Human Plasma Proteome Draft of 2017: Building on the Human Plasma PeptideAtlas from Mass Spectrometry and Complementary Assays. J. Proteome Res. 2017;16:4299–4310. doi: 10.1021/acs.jproteome.7b00467. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous