A Non-Replicative Role of the 3' Terminal Sequence of the Dengue Virus Genome in Membranous Replication Organelle Formation
- PMID: 32640225
- PMCID: PMC7351112
- DOI: 10.1016/j.celrep.2020.107859
A Non-Replicative Role of the 3' Terminal Sequence of the Dengue Virus Genome in Membranous Replication Organelle Formation
Abstract
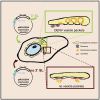
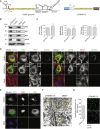
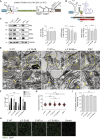
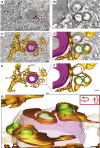
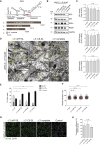
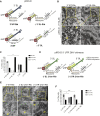
Dengue virus (DENV) and Zika virus (ZIKV), members of the Flavivirus genus, rearrange endoplasmic reticulum membranes to induce invaginations known as vesicle packets (VPs), which are the assumed sites for viral RNA replication. Mechanistic information on VP biogenesis has so far been difficult to attain due to the necessity of studying their formation under conditions of viral replication, where perturbations reducing replication will inevitably impact VP formation. Here, we report a replication-independent expression system, designated pIRO (plasmid-induced replication organelle formation) that induces bona fide DENV and ZIKV VPs that are morphologically indistinguishable from those in infected cells. Using this system, we demonstrate that sequences in the 3' terminal RNA region of the DENV, but not the ZIKV genome, contribute to VP formation in a non-replicative manner. These results validate the pIRO system that opens avenues for mechanistically dissecting virus replication from membrane reorganization.
Keywords: flavivirus; membrane invagination; membranous organelle; organelle biogenesis; replication complex; replication organelle; vesicle packet; viral replicase.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures


Similar articles
-
Determinants in Nonstructural Protein 4A of Dengue Virus Required for RNA Replication and Replication Organelle Biogenesis.J Virol. 2021 Oct 13;95(21):e0131021. doi: 10.1128/JVI.01310-21. Epub 2021 Aug 11. J Virol. 2021. PMID: 34379504 Free PMC article.
-
A novel interaction between dengue virus nonstructural protein 1 and the NS4A-2K-4B precursor is required for viral RNA replication but not for formation of the membranous replication organelle.PLoS Pathog. 2019 May 9;15(5):e1007736. doi: 10.1371/journal.ppat.1007736. eCollection 2019 May. PLoS Pathog. 2019. PMID: 31071189 Free PMC article.
-
Genome-Wide CRISPR Screen Identifies RACK1 as a Critical Host Factor for Flavivirus Replication.J Virol. 2021 Nov 23;95(24):e0059621. doi: 10.1128/JVI.00596-21. Epub 2021 Sep 29. J Virol. 2021. PMID: 34586867 Free PMC article.
-
Control of dengue virus translation and replication.Curr Top Microbiol Immunol. 2010;338:15-34. doi: 10.1007/978-3-642-02215-9_2. Curr Top Microbiol Immunol. 2010. PMID: 19802575 Review.
-
Functional RNA elements in the dengue virus genome.Viruses. 2011 Sep;3(9):1739-56. doi: 10.3390/v3091739. Epub 2011 Sep 15. Viruses. 2011. PMID: 21994804 Free PMC article. Review.
Cited by
-
Flavivirus nonstructural proteins and replication complexes as antiviral drug targets.Curr Opin Virol. 2023 Apr;59:101305. doi: 10.1016/j.coviro.2023.101305. Epub 2023 Mar 2. Curr Opin Virol. 2023. PMID: 36870091 Free PMC article. Review.
-
A Versatile Reporter System To Monitor Virus-Infected Cells and Its Application to Dengue Virus and SARS-CoV-2.J Virol. 2021 Jan 28;95(4):e01715-20. doi: 10.1128/JVI.01715-20. Print 2021 Jan 28. J Virol. 2021. PMID: 33257477 Free PMC article.
-
Molecular Insights into the Flavivirus Replication Complex.Viruses. 2021 May 21;13(6):956. doi: 10.3390/v13060956. Viruses. 2021. PMID: 34064113 Free PMC article. Review.
-
Pan-serotype dengue virus inhibitor JNJ-A07 targets NS4A-2K-NS4B interaction with NS2B/NS3 and blocks replication organelle formation.Nat Commun. 2024 Jul 19;15(1):6080. doi: 10.1038/s41467-024-50437-3. Nat Commun. 2024. PMID: 39030239 Free PMC article.
-
N-terminal acetyltransferase 6 facilitates enterovirus 71 replication by regulating PI4KB expression and replication organelle biogenesis.J Virol. 2024 Feb 20;98(2):e0174923. doi: 10.1128/jvi.01749-23. Epub 2024 Jan 8. J Virol. 2024. PMID: 38189249 Free PMC article.
References
-
- Aktepe T.E., Liebscher S., Prier J.E., Simmons C.P., Mackenzie J.M. The Host Protein Reticulon 3.1A Is Utilized by Flaviviruses to Facilitate Membrane Remodelling. Cell Rep. 2017;21:1639–1654. - PubMed
-
- Alvarez D.E., De Lella Ezcurra A.L., Fucito S., Gamarnik A.V. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005;339:200–212. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

