Canonical and Noncanonical Autophagy as Potential Targets for COVID-19
- PMID: 32635598
- PMCID: PMC7408018
- DOI: 10.3390/cells9071619
Canonical and Noncanonical Autophagy as Potential Targets for COVID-19
Abstract
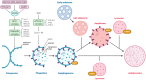
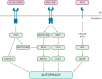
The SARS-CoV-2 pandemic necessitates a review of the molecular mechanisms underlying cellular infection by coronaviruses, in order to identify potential therapeutic targets against the associated new disease (COVID-19). Previous studies on its counterparts prove a complex and concomitant interaction between coronaviruses and autophagy. The precise manipulation of this pathway allows these viruses to exploit the autophagy molecular machinery while avoiding its protective apoptotic drift and cellular innate immune responses. In turn, the maneuverability margins of such hijacking appear to be so narrow that the modulation of the autophagy, regardless of whether using inducers or inhibitors (many of which are FDA-approved for the treatment of other diseases), is usually detrimental to viral replication, including SARS-CoV-2. Recent discoveries indicate that these interactions stretch into the still poorly explored noncanonical autophagy pathway, which might play a substantial role in coronavirus replication. Still, some potential therapeutic targets within this pathway, such as RAB9 and its interacting proteins, look promising considering current knowledge. Thus, the combinatory treatment of COVID-19 with drugs affecting both canonical and noncanonical autophagy pathways may be a turning point in the fight against this and other viral infections, which may also imply beneficial prospects of long-term protection.
Keywords: COVID-19; SARS-CoV-2; antiviral; autophagy; canonical autophagy; coronavirus; noncanonical autophagy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
A pharmacological perspective of chloroquine in SARS-CoV-2 infection: An old drug for the fight against a new coronavirus?Int J Antimicrob Agents. 2020 Sep;56(3):106078. doi: 10.1016/j.ijantimicag.2020.106078. Epub 2020 Jul 4. Int J Antimicrob Agents. 2020. PMID: 32629115 Free PMC article. Review.
-
3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.Sci Transl Med. 2020 Aug 19;12(557):eabc5332. doi: 10.1126/scitranslmed.abc5332. Epub 2020 Aug 3. Sci Transl Med. 2020. PMID: 32747425 Free PMC article.
-
Targeting the Endocytic Pathway and Autophagy Process as a Novel Therapeutic Strategy in COVID-19.Int J Biol Sci. 2020 Mar 15;16(10):1724-1731. doi: 10.7150/ijbs.45498. eCollection 2020. Int J Biol Sci. 2020. PMID: 32226290 Free PMC article. Review.
-
Potential RNA-dependent RNA polymerase inhibitors as prospective therapeutics against SARS-CoV-2.J Med Microbiol. 2020 Jun;69(6):864-873. doi: 10.1099/jmm.0.001203. Epub 2020 May 29. J Med Microbiol. 2020. PMID: 32469301 Free PMC article.
-
COVID-19/SARS-CoV-2 Infection: Lysosomes and Lysosomotropism Implicate New Treatment Strategies and Personal Risks.Int J Mol Sci. 2020 Jul 13;21(14):4953. doi: 10.3390/ijms21144953. Int J Mol Sci. 2020. PMID: 32668803 Free PMC article. Review.
Cited by
-
Alarmins, COVID-19 and comorbidities.Ann Med. 2021 Dec;53(1):777-785. doi: 10.1080/07853890.2021.1921252. Ann Med. 2021. PMID: 34042528 Free PMC article. Review.
-
The relationship between autophagy and respiratory viruses.Arch Microbiol. 2024 Mar 4;206(4):136. doi: 10.1007/s00203-024-03838-3. Arch Microbiol. 2024. PMID: 38436746 Review.
-
Vascular underpinning of COVID-19.Open Biol. 2020 Aug;10(8):200208. doi: 10.1098/rsob.200208. Epub 2020 Aug 27. Open Biol. 2020. PMID: 32847471 Free PMC article.
-
The potential link between Covid-19 and multiple myeloma: A new saga.Immun Inflamm Dis. 2022 Dec;10(12):e701. doi: 10.1002/iid3.701. Immun Inflamm Dis. 2022. PMID: 36444620 Free PMC article. Review.
-
Neutrophil autophagy and NETosis in COVID-19: perspectives.Autophagy. 2023 Mar;19(3):758-767. doi: 10.1080/15548627.2022.2099206. Epub 2022 Aug 11. Autophagy. 2023. PMID: 35951555 Free PMC article. Review.
References
-
- WHO . International Health Regulations (2005) World Health Organization; Geneva, Switzerland: 2008.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

