Impact of Losing hRpn13 Pru or UCHL5 on Proteasome Clearance of Ubiquitinated Proteins and RA190 Cytotoxicity
- PMID: 32631902
- PMCID: PMC7459265
- DOI: 10.1128/MCB.00122-20
Impact of Losing hRpn13 Pru or UCHL5 on Proteasome Clearance of Ubiquitinated Proteins and RA190 Cytotoxicity
Abstract
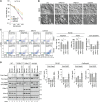
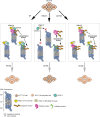
hRpn13/ADRM1 links substrate recruitment with deubiquitination at the proteasome through its proteasome- and ubiquitin-binding Pru domain and DEUBAD domain, which binds and activates deubiquitinating enzyme (DUB) UCHL5/Uch37. Here, we edit the HCT116 colorectal cancer cell line to delete part of the hRpn13 Pru, producing cells that express truncated hRpn13 (trRpn13), which is competent for UCHL5 binding but defective for proteasome interaction. trRpn13 cells demonstrate reduced levels of proteasome-bound ubiquitinated proteins, indicating that the loss of hRpn13 function at proteasomes cannot be fully compensated for by the two other dedicated substrate receptors (hRpn1 and hRpn10). Previous studies indicated that the loss of full-length hRpn13 causes a corresponding reduction of UCHL5. We find UCHL5 levels unaltered in trRpn13 cells, but hRpn11 is elevated in ΔhRpn13 and trRpn13 cells, perhaps from cell stress. Despite the ∼90 DUBs in human cells, including two others in addition to UCHL5 at the proteasome, we found deletion of UCHL5 from HCT116 cells to cause increased levels of ubiquitinated proteins in whole-cell extract and at proteasomes, suggesting that UCHL5 activity cannot be fully assumed by other DUBs. We also report anticancer molecule RA190, which binds covalently to hRpn13 and UCHL5, to require hRpn13 Pru and not UCHL5 for cytotoxicity.
Keywords: ADRM1; Adrm1; RA190; Rpn13; UCHL5; Uch37; apoptosis; cell cycle progression; cell viability; proteasome; ubiquitin.
Figures




Similar articles
-
Structure of the Rpn13-Rpn2 complex provides insights for Rpn13 and Uch37 as anticancer targets.Nat Commun. 2017 Jun 9;8:15540. doi: 10.1038/ncomms15540. Nat Commun. 2017. PMID: 28598414 Free PMC article.
-
The Proteasome Ubiquitin Receptor hRpn13 and Its Interacting Deubiquitinating Enzyme Uch37 Are Required for Proper Cell Cycle Progression.J Biol Chem. 2016 Apr 15;291(16):8773-83. doi: 10.1074/jbc.M115.694588. Epub 2016 Feb 23. J Biol Chem. 2016. PMID: 26907685 Free PMC article.
-
A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes.EMBO J. 2006 Oct 4;25(19):4524-36. doi: 10.1038/sj.emboj.7601338. Epub 2006 Sep 21. EMBO J. 2006. PMID: 16990800 Free PMC article.
-
Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes.Mol Cell Proteomics. 2011 May;10(5):R110.003871. doi: 10.1074/mcp.R110.003871. Epub 2010 Sep 7. Mol Cell Proteomics. 2011. PMID: 20823120 Free PMC article. Review.
-
Role of the ubiquitin-proteasome system in brain ischemia: friend or foe?Prog Neurobiol. 2014 Jan;112:50-69. doi: 10.1016/j.pneurobio.2013.10.003. Epub 2013 Oct 22. Prog Neurobiol. 2014. PMID: 24157661 Review.
Cited by
-
Potential roles of UCH family deubiquitinases in tumorigenesis and chemical inhibitors developed against them.Am J Cancer Res. 2024 Jun 15;14(6):2666-2694. doi: 10.62347/OEGE2648. eCollection 2024. Am J Cancer Res. 2024. PMID: 39005671 Free PMC article. Review.
-
Proteasome-Bound UCH37/UCHL5 Debranches Ubiquitin Chains to Promote Degradation.Mol Cell. 2020 Dec 3;80(5):796-809.e9. doi: 10.1016/j.molcel.2020.10.017. Epub 2020 Nov 5. Mol Cell. 2020. PMID: 33156996 Free PMC article.
-
Development and anticancer properties of Up284, a spirocyclic candidate ADRM1/RPN13 inhibitor.PLoS One. 2023 Jun 14;18(6):e0285221. doi: 10.1371/journal.pone.0285221. eCollection 2023. PLoS One. 2023. PMID: 37315065 Free PMC article.
-
A structure-based designed small molecule depletes hRpn13Pru and a select group of KEN box proteins.Nat Commun. 2024 Mar 20;15(1):2485. doi: 10.1038/s41467-024-46644-7. Nat Commun. 2024. PMID: 38509117 Free PMC article.
-
hRpn13 shapes the proteome and transcriptome through epigenetic factors HDAC8, PADI4, and transcription factor NF-κB p50.Mol Cell. 2024 Feb 1;84(3):522-537.e8. doi: 10.1016/j.molcel.2023.11.035. Epub 2023 Dec 26. Mol Cell. 2024. PMID: 38151017 Free PMC article.
References
-
- Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee BH, Shi Y, Zhang N, de Poot SA, Tuebing F, Sun S, Vannoy J, Tarasov SG, Engen JR, Finley D, Walters KJ. 2016. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 351:aad9421. doi:10.1126/science.aad9421. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
