Chemical Class Prediction of Unknown Biomolecules Using Ion Mobility-Mass Spectrometry and Machine Learning: Supervised Inference of Feature Taxonomy from Ensemble Randomization
- PMID: 32628452
- PMCID: PMC9374365
- DOI: 10.1021/acs.analchem.0c02137
Chemical Class Prediction of Unknown Biomolecules Using Ion Mobility-Mass Spectrometry and Machine Learning: Supervised Inference of Feature Taxonomy from Ensemble Randomization
Abstract
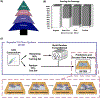
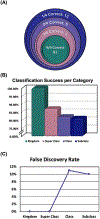
This work presents a machine learning algorithm referred to as the supervised inference of feature taxonomy from ensemble randomization (SIFTER), which supports the identification of features derived from untargeted ion mobility-mass spectrometry (IM-MS) experiments. SIFTER utilizes random forest machine learning on three analytical measurements derived from IM-MS (collision cross section, CCS), mass-to-charge (m/z), and mass defect (Δm) to classify unknown features into a taxonomy of chemical kingdom, super class, class, and subclass. Each of these classifications is assigned a calculated probability as well as alternate classifications with associated probabilities. After optimization, SIFTER was tested against a set of molecules not used in the training set. The average success rate in classifying all four taxonomy categories correctly was found to be >99%. Analysis of molecular features detected from a complex biological matrix and not used in the training set yielded a lower success rate where all four categories were correctly predicted for ∼80% of the compounds. This decline in performance is in part due to incompleteness of the training set across all potential taxonomic categories, but also resulting from a nearest-neighbor bias in the random forest algorithm. Ongoing efforts are focused on improving the class prediction accuracy of SIFTER through expansion of empirical data sets used for training as well as improvements to the core algorithm.
Figures



Similar articles
-
Unknown Metabolite Identification Using Machine Learning Collision Cross-Section Prediction and Tandem Mass Spectrometry.Anal Chem. 2023 Jan 17;95(2):1047-1056. doi: 10.1021/acs.analchem.2c03749. Epub 2023 Jan 3. Anal Chem. 2023. PMID: 36595469 Free PMC article.
-
Large-Scale Prediction of Collision Cross-Section Values for Metabolites in Ion Mobility-Mass Spectrometry.Anal Chem. 2016 Nov 15;88(22):11084-11091. doi: 10.1021/acs.analchem.6b03091. Epub 2016 Nov 1. Anal Chem. 2016. PMID: 27768289
-
Collision Cross Section Prediction with Molecular Fingerprint Using Machine Learning.Molecules. 2022 Sep 29;27(19):6424. doi: 10.3390/molecules27196424. Molecules. 2022. PMID: 36234961 Free PMC article.
-
Determination of drugs and drug metabolites by ion mobility-mass spectrometry: A review.Anal Chim Acta. 2021 Apr 15;1154:338270. doi: 10.1016/j.aca.2021.338270. Epub 2021 Feb 2. Anal Chim Acta. 2021. PMID: 33736803 Review.
-
Advancing the large-scale CCS database for metabolomics and lipidomics at the machine-learning era.Curr Opin Chem Biol. 2018 Feb;42:34-41. doi: 10.1016/j.cbpa.2017.10.033. Epub 2017 Nov 12. Curr Opin Chem Biol. 2018. PMID: 29136580 Review.
Cited by
-
CCS Predictor 2.0: An Open-Source Jupyter Notebook Tool for Filtering Out False Positives in Metabolomics.Anal Chem. 2022 Dec 20;94(50):17456-17466. doi: 10.1021/acs.analchem.2c03491. Epub 2022 Dec 6. Anal Chem. 2022. PMID: 36473057 Free PMC article.
-
Collision Cross Section Prediction Based on Machine Learning.Molecules. 2023 May 12;28(10):4050. doi: 10.3390/molecules28104050. Molecules. 2023. PMID: 37241791 Free PMC article. Review.
-
Improving confidence in lipidomic annotations by incorporating empirical ion mobility regression analysis and chemical class prediction.Bioinformatics. 2022 May 13;38(10):2872-2879. doi: 10.1093/bioinformatics/btac197. Bioinformatics. 2022. PMID: 35561172 Free PMC article.
-
Improved analysis of derivatized steroid hormone isomers using ion mobility-mass spectrometry (IM-MS).Anal Bioanal Chem. 2023 Nov;415(27):6757-6769. doi: 10.1007/s00216-023-04953-8. Epub 2023 Sep 23. Anal Bioanal Chem. 2023. PMID: 37740752
-
Multidimensional Separations of Intact Phase II Steroid Metabolites Utilizing LC-Ion Mobility-HRMS.Anal Chem. 2021 Aug 10;93(31):10990-10998. doi: 10.1021/acs.analchem.1c02163. Epub 2021 Jul 28. Anal Chem. 2021. PMID: 34319704 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

