Conserved metabolite regulation of stress granule assembly via AdoMet
- PMID: 32609300
- PMCID: PMC7401819
- DOI: 10.1083/jcb.201904141
Conserved metabolite regulation of stress granule assembly via AdoMet
Abstract
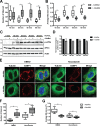
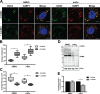
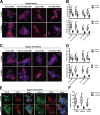
Stress granules (SGs) are evolutionarily conserved condensates of ribonucleoproteins that assemble in response to metabolic stresses. Because aberrant SG formation is associated with amyotrophic lateral sclerosis (ALS), understanding the connection between metabolic activity and SG composition can provide therapeutic insights into neurodegeneration. Here, we identify 17 metabolic enzymes recruited to yeast SGs in response to physiological growth stress. Furthermore, the product of one of these enzymes, AdoMet, is a regulator of SG assembly and composition. Decreases in AdoMet levels increase SG formation, while chronic elevation of AdoMet produces SG remnants lacking proteins associated with the 5' end of transcripts. Interestingly, acute elevation of AdoMet blocks SG formation in yeast and motor neurons. Treatment of ALS-derived motor neurons with AdoMet also suppresses the formation of TDP-43-positive SGs, a hallmark of ALS. Together, these results argue that AdoMet is an evolutionarily conserved regulator of SG composition and assembly with therapeutic potential in neurodegeneration.
© 2020 Begovich et al.
Figures



Similar articles
-
Stress granules in neurodegeneration--lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma.FEBS J. 2013 Sep;280(18):4348-70. doi: 10.1111/febs.12287. Epub 2013 May 9. FEBS J. 2013. PMID: 23587065 Review.
-
Small-Molecule Modulation of TDP-43 Recruitment to Stress Granules Prevents Persistent TDP-43 Accumulation in ALS/FTD.Neuron. 2019 Sep 4;103(5):802-819.e11. doi: 10.1016/j.neuron.2019.05.048. Epub 2019 Jul 1. Neuron. 2019. PMID: 31272829 Free PMC article.
-
Pur-alpha regulates cytoplasmic stress granule dynamics and ameliorates FUS toxicity.Acta Neuropathol. 2016 Apr;131(4):605-20. doi: 10.1007/s00401-015-1530-0. Epub 2016 Jan 4. Acta Neuropathol. 2016. PMID: 26728149 Free PMC article.
-
Spatiotemporal Proteomic Analysis of Stress Granule Disassembly Using APEX Reveals Regulation by SUMOylation and Links to ALS Pathogenesis.Mol Cell. 2020 Dec 3;80(5):876-891.e6. doi: 10.1016/j.molcel.2020.10.032. Epub 2020 Nov 19. Mol Cell. 2020. PMID: 33217318 Free PMC article.
-
Stress Granule Homeostasis, Aberrant Phase Transition, and Amyotrophic Lateral Sclerosis.ACS Chem Neurosci. 2022 Aug 17;13(16):2356-2370. doi: 10.1021/acschemneuro.2c00262. Epub 2022 Jul 29. ACS Chem Neurosci. 2022. PMID: 35905138 Review.
Cited by
-
The Role of Methionine Residues in the Regulation of Liquid-Liquid Phase Separation.Biomolecules. 2021 Aug 21;11(8):1248. doi: 10.3390/biom11081248. Biomolecules. 2021. PMID: 34439914 Free PMC article. Review.
-
Stress-related biomolecular condensates in plants.Plant Cell. 2023 Sep 1;35(9):3187-3204. doi: 10.1093/plcell/koad127. Plant Cell. 2023. PMID: 37162152 Free PMC article.
-
Reversible amyloids of pyruvate kinase couple cell metabolism and stress granule disassembly.Nat Cell Biol. 2021 Oct;23(10):1085-1094. doi: 10.1038/s41556-021-00760-4. Epub 2021 Oct 6. Nat Cell Biol. 2021. PMID: 34616026 Free PMC article.
-
Plant Stress Granules: Trends and Beyond.Front Plant Sci. 2021 Aug 9;12:722643. doi: 10.3389/fpls.2021.722643. eCollection 2021. Front Plant Sci. 2021. PMID: 34434210 Free PMC article. Review.
-
Electrochemical Measurements Reveal Reactive Oxygen Species in Stress Granules*.Angew Chem Int Ed Engl. 2021 Jul 5;60(28):15302-15306. doi: 10.1002/anie.202104308. Epub 2021 Jun 9. Angew Chem Int Ed Engl. 2021. PMID: 33876544 Free PMC article.
References
-
- Aguilera-Gomez A., Zacharogianni M., van Oorschot M.M., Genau H., Grond R., Veenendaal T., Sinsimer K.S., Gavis E.R., Behrends C., and Rabouille C.. 2017. Phospho-Rasputin stabilization by Sec16 is required for stress granule formation upon amino acid starvation. Cell Rep. 20:935–948. 10.1016/j.celrep.2017.06.042 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

