A novel FRET peptide assay reveals efficient Helicobacter pylori HtrA inhibition through zinc and copper binding
- PMID: 32601479
- PMCID: PMC7324608
- DOI: 10.1038/s41598-020-67578-2
A novel FRET peptide assay reveals efficient Helicobacter pylori HtrA inhibition through zinc and copper binding
Abstract
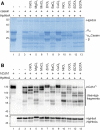
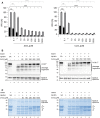
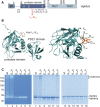
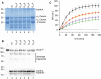
Helicobacter pylori (H. pylori) secretes the chaperone and serine protease high temperature requirement A (HtrA) that cleaves gastric epithelial cell surface proteins to disrupt the epithelial integrity and barrier function. First inhibitory lead structures have demonstrated the essential role of HtrA in H. pylori physiology and pathogenesis. Comprehensive drug discovery techniques allowing high-throughput screening are now required to develop effective compounds. Here, we designed a novel fluorescence resonance energy transfer (FRET) peptide derived from a gel-based label-free proteomic approach (direct in-gel profiling of protease specificity) as a valuable substrate for H. pylori HtrA. Since serine proteases are often sensitive to metal ions, we investigated the influence of different divalent ions on the activity of HtrA. We identified Zn++ and Cu++ ions as inhibitors of H. pylori HtrA activity, as monitored by in vitro cleavage experiments using casein or E-cadherin as substrates and in the FRET peptide assay. Putative binding sites for Zn++ and Cu++ were then analyzed in thermal shift and microscale thermophoresis assays. The findings of this study will contribute to the development of novel metal ion-dependent protease inhibitors, which might help to fight bacterial infections.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Identification of E-cadherin signature motifs functioning as cleavage sites for Helicobacter pylori HtrA.Sci Rep. 2016 Mar 17;6:23264. doi: 10.1038/srep23264. Sci Rep. 2016. PMID: 26983597 Free PMC article.
-
Chaperone activity of serine protease HtrA of Helicobacter pylori as a crucial survival factor under stress conditions.Cell Commun Signal. 2019 Dec 3;17(1):161. doi: 10.1186/s12964-019-0481-9. Cell Commun Signal. 2019. PMID: 31796064 Free PMC article.
-
Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion.EMBO Rep. 2010 Oct;11(10):798-804. doi: 10.1038/embor.2010.114. Epub 2010 Sep 3. EMBO Rep. 2010. PMID: 20814423 Free PMC article.
-
Proteolysis in Helicobacter pylori-Induced Gastric Cancer.Toxins (Basel). 2017 Apr 11;9(4):134. doi: 10.3390/toxins9040134. Toxins (Basel). 2017. PMID: 28398251 Free PMC article. Review.
-
Two remarkable serine/leucine polymorphisms in Helicobacter pylori: functional importance for serine protease HtrA and adhesin BabA.Cell Commun Signal. 2024 May 2;22(1):250. doi: 10.1186/s12964-024-01635-5. Cell Commun Signal. 2024. PMID: 38698410 Free PMC article. Review.
Cited by
-
Identification of Desmoglein-2 as a novel target of Helicobacter pylori HtrA in epithelial cells.Cell Commun Signal. 2021 Nov 6;19(1):108. doi: 10.1186/s12964-021-00788-x. Cell Commun Signal. 2021. PMID: 34742300 Free PMC article.
-
E-Cadherin Orthologues as Substrates for the Serine Protease High Temperature Requirement A (HtrA).Biomolecules. 2022 Feb 24;12(3):356. doi: 10.3390/biom12030356. Biomolecules. 2022. PMID: 35327548 Free PMC article.
-
Targeting Peptides: The New Generation of Targeted Drug Delivery Systems.Pharmaceutics. 2023 Jun 3;15(6):1648. doi: 10.3390/pharmaceutics15061648. Pharmaceutics. 2023. PMID: 37376097 Free PMC article. Review.
-
Structural basis of substrate recognition and allosteric activation of the proapoptotic mitochondrial HtrA2 protease.Nat Commun. 2024 May 30;15(1):4592. doi: 10.1038/s41467-024-48997-5. Nat Commun. 2024. PMID: 38816423 Free PMC article.
-
L-tryptophan and copper interactions linked to reduced colibactin genotoxicity in pks+ Escherichia coli.mSystems. 2024 Oct 22;9(10):e0099224. doi: 10.1128/msystems.00992-24. Epub 2024 Sep 12. mSystems. 2024. PMID: 39264195 Free PMC article.
References
-
- Global Burden of Disease Cancer Collaboration Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

