Candida auris Phenotypic Heterogeneity Determines Pathogenicity In Vitro
- PMID: 32581078
- PMCID: PMC7316489
- DOI: 10.1128/mSphere.00371-20
Candida auris Phenotypic Heterogeneity Determines Pathogenicity In Vitro
Abstract
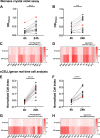
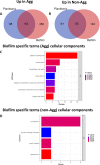
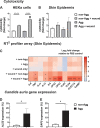
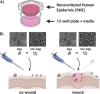
Candida auris is an enigmatic yeast that provides substantial global risk in health care facilities and intensive care units. A unique phenotype exhibited by certain isolates of C. auris is their ability to form small clusters of cells known as aggregates, which have been to a limited extent described in the context of pathogenic traits. In this study, we screened several nonaggregative and aggregative C. auris isolates for biofilm formation, where we observed a level of heterogeneity among the different phenotypes. Next, we utilized an RNA sequencing approach to investigate the transcriptional responses during biofilm formation of a nonaggregative and aggregative isolate of the initial pool. Observations from these analyses indicate unique transcriptional profiles in the two isolates, with several genes identified relating to proteins involved in adhesion and invasion of the host in other fungal species. From these findings, we investigated for the first time the fungal recognition and inflammatory responses of a three-dimensional skin epithelial model to these isolates. In these models, a wound was induced to mimic a portal of entry for C. auris We show that both phenotypes elicited minimal response in the model minus induction of the wound, yet in the wounded tissue, both phenotypes induced a greater response, with the aggregative isolate more proinflammatory. This capacity of aggregative C. auris biofilms to generate such responses in the wounded skin highlights how this opportunistic yeast is a high risk within the intensive care environment where susceptible patients have multiple indwelling lines.IMPORTANCECandida auris has recently emerged as an important cause of concern within health care environments due to its ability to persist and tolerate commonly used antiseptics and disinfectants, particularly when attached to a surface (biofilms). This yeast is able to colonize and subsequently infect patients, particularly those that are critically ill or immunosuppressed, which may result in death. We have undertaken analysis on two different phenotypic types of this yeast, using molecular and immunological tools to determine whether either of these has a greater ability to cause serious infections. We describe that both isolates exhibit largely different transcriptional profiles during biofilm development. Finally, we show that the inability to form small aggregates (or clusters) of cells has an adverse effect on the organism's immunostimulatory properties, suggesting that the nonaggregative phenotype may exhibit a certain level of immune evasion.
Keywords: Candida auris; aggregate; heterogeneity; host-pathogen interactions; in vitro skin model.
Copyright © 2020 Brown et al.
Figures
Similar articles
-
Biofilm-Forming Capability of Highly Virulent, Multidrug-Resistant Candida auris.Emerg Infect Dis. 2017 Feb;23(2):328-331. doi: 10.3201/eid2302.161320. Emerg Infect Dis. 2017. PMID: 28098553 Free PMC article.
-
Comparative Evaluations of the Pathogenesis of Candida auris Phenotypes and Candida albicans Using Clinically Relevant Murine Models of Infections.mSphere. 2020 Aug 5;5(4):e00760-20. doi: 10.1128/mSphere.00760-20. mSphere. 2020. PMID: 32759340 Free PMC article.
-
Biofilm formation by Candida auris isolated from colonising sites and candidemia cases.Mycoses. 2019 Aug;62(8):706-709. doi: 10.1111/myc.12947. Epub 2019 Jun 9. Mycoses. 2019. PMID: 31132181
-
What do we know about the biology of the emerging fungal pathogen of humans Candida auris?Microbiol Res. 2021 Jan;242:126621. doi: 10.1016/j.micres.2020.126621. Epub 2020 Oct 9. Microbiol Res. 2021. PMID: 33096325 Review.
-
Intra-clade Heterogeneity in Candida auris: Risk of Management.Curr Microbiol. 2023 Jul 24;80(9):295. doi: 10.1007/s00284-023-03416-8. Curr Microbiol. 2023. PMID: 37486431 Review.
Cited by
-
Relatedness and the evolution of mechanisms to divide labor in microorganisms.Ecol Evol. 2021 Oct 8;11(21):14475-14489. doi: 10.1002/ece3.8067. eCollection 2021 Nov. Ecol Evol. 2021. PMID: 34765120 Free PMC article.
-
Yeast and filamentous Candida auris stimulate distinct immune responses in the skin.mSphere. 2024 Jul 30;9(7):e0005524. doi: 10.1128/msphere.00055-24. Epub 2024 Jun 21. mSphere. 2024. PMID: 38904381 Free PMC article.
-
Synergistic Antifungal Interaction between Pseudomonas aeruginosa LV Strain Metabolites and Biogenic Silver Nanoparticles against Candida auris.Antibiotics (Basel). 2023 May 6;12(5):861. doi: 10.3390/antibiotics12050861. Antibiotics (Basel). 2023. PMID: 37237764 Free PMC article.
-
Cell aggregation mediated by ACE2 deletion in Candida auris modulates fungal colonization and host immune responses in the skin.mSphere. 2024 Nov 21;9(11):e0073424. doi: 10.1128/msphere.00734-24. Epub 2024 Oct 30. mSphere. 2024. PMID: 39475280 Free PMC article.
-
Assessing the Bioactive Profile of Antifungal-Loaded Calcium Sulfate against Fungal Biofilms.Antimicrob Agents Chemother. 2021 May 18;65(6):e02551-20. doi: 10.1128/AAC.02551-20. Print 2021 May 18. Antimicrob Agents Chemother. 2021. PMID: 33753336 Free PMC article.
References
-
- Kim MN, Shin JH, Sung H, Lee K, Kim EC, Ryoo N, Lee JS, Jung SI, Park KH, Kee SJ, Kim SH, Shin MG, Suh SP, Ryang DW. 2009. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis 48:e57–e61. doi:10.1086/597108. - DOI - PubMed
-
- Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi:10.1093/cid/ciw691. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
