Deciphering the TCR Repertoire to Solve the COVID-19 Mystery
- PMID: 32576386
- PMCID: PMC7305739
- DOI: 10.1016/j.tips.2020.06.001
Deciphering the TCR Repertoire to Solve the COVID-19 Mystery
Abstract
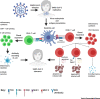
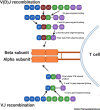
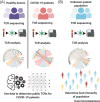
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected several millions and killed more than quarter of a million worldwide to date. Important questions have remained unanswered: why some patients develop severe disease, while others do not; and what roles do genetic variabilities play in the individual immune response to this viral infection. Here, we discuss the critical role T cells play in the orchestration of the antiviral response underlying the pathogenesis of the disease, COVID-19. We highlight the scientific rationale for comprehensive and longitudinal TCR analyses in COVID-19 and reason that analyzing TCR repertoire in COVID-19 patients would reveal important findings that may explain the outcome disparity observed in these patients. Finally, we provide a framework describing the different strategies, the advantages, and the challenges involved in obtaining useful TCR repertoire data to advance our fight against COVID-19.
Keywords: COVID-19; SARS-CoV-2; T cell receptors; TCR repertoire; TCR-seq.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals.Signal Transduct Target Ther. 2020 Aug 14;5(1):156. doi: 10.1038/s41392-020-00263-y. Signal Transduct Target Ther. 2020. PMID: 32796814 Free PMC article.
-
Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25254-25262. doi: 10.1073/pnas.2010722117. Epub 2020 Sep 28. Proc Natl Acad Sci U S A. 2020. PMID: 32989130 Free PMC article.
-
Profiling of T Cell Repertoire in SARS-CoV-2-Infected COVID-19 Patients Between Mild Disease and Pneumonia.J Clin Immunol. 2021 Aug;41(6):1131-1145. doi: 10.1007/s10875-021-01045-z. Epub 2021 May 5. J Clin Immunol. 2021. PMID: 33950324 Free PMC article.
-
Deciphering the Role of Host Genetics in Susceptibility to Severe COVID-19.Front Immunol. 2020 Jun 30;11:1606. doi: 10.3389/fimmu.2020.01606. eCollection 2020. Front Immunol. 2020. PMID: 32695122 Free PMC article. Review.
-
[Adaptive immunity against SARS-CoV-2].Med Sci (Paris). 2020 Oct;36(10):908-913. doi: 10.1051/medsci/2020168. Epub 2020 Sep 22. Med Sci (Paris). 2020. PMID: 32960167 Review. French.
Cited by
-
The molecular mechanisms of CD8+ T cell responses to SARS-CoV-2 infection mediated by TCR-pMHC interactions.Front Immunol. 2024 Oct 10;15:1468456. doi: 10.3389/fimmu.2024.1468456. eCollection 2024. Front Immunol. 2024. PMID: 39450171 Free PMC article. Review.
-
Immune responses during COVID-19 infection.Oncoimmunology. 2020 Aug 25;9(1):1807836. doi: 10.1080/2162402X.2020.1807836. Oncoimmunology. 2020. PMID: 32939324 Free PMC article. Review.
-
Molecular signature of postmortem lung tissue from COVID-19 patients suggests distinct trajectories driving mortality.Dis Model Mech. 2022 May 1;15(5):dmm049572. doi: 10.1242/dmm.049572. Epub 2022 Jun 6. Dis Model Mech. 2022. PMID: 35438176 Free PMC article.
-
CoVITEST: A Fast and Reliable Method to Monitor Anti-SARS-CoV-2 Specific T Cells From Whole Blood.Front Immunol. 2022 Jul 5;13:848586. doi: 10.3389/fimmu.2022.848586. eCollection 2022. Front Immunol. 2022. PMID: 35865538 Free PMC article.
-
Healing Field: Using Alternating Electric Fields to Prevent Cytokine Storm by Suppressing Clonal Expansion of the Activated Lymphocytes in the Blood Sample of the COVID-19 Patients.Front Bioeng Biotechnol. 2022 Jun 2;10:850571. doi: 10.3389/fbioe.2022.850571. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35721862 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

