Epigenetic priming by Dppa2 and 4 in pluripotency facilitates multi-lineage commitment
- PMID: 32572255
- PMCID: PMC7614975
- DOI: 10.1038/s41594-020-0443-3
Epigenetic priming by Dppa2 and 4 in pluripotency facilitates multi-lineage commitment
Abstract
How the epigenetic landscape is established in development is still being elucidated. Here, we uncover developmental pluripotency associated 2 and 4 (DPPA2/4) as epigenetic priming factors that establish a permissive epigenetic landscape at a subset of developmentally important bivalent promoters characterized by low expression and poised RNA-polymerase. Differentiation assays reveal that Dppa2/4 double knockout mouse embryonic stem cells fail to exit pluripotency and differentiate efficiently. DPPA2/4 bind both H3K4me3-marked and bivalent gene promoters and associate with COMPASS- and Polycomb-bound chromatin. Comparing knockout and inducible knockdown systems, we find that acute depletion of DPPA2/4 results in rapid loss of H3K4me3 from key bivalent genes, while H3K27me3 is initially more stable but lost following extended culture. Consequently, upon DPPA2/4 depletion, these promoters gain DNA methylation and are unable to be activated upon differentiation. Our findings uncover a novel epigenetic priming mechanism at developmental promoters, poising them for future lineage-specific activation.
Conflict of interest statement
W.R. is a consultant and shareholder of Cambridge Epigenetix. The remaining authors declare no competing financial interests.
Figures

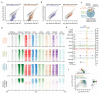
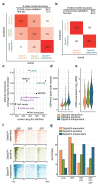


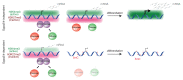
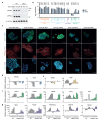




Comment in
-
Am-bivalency towards DNA methylation.Nat Rev Mol Cell Biol. 2020 Sep;21(9):497. doi: 10.1038/s41580-020-0274-4. Nat Rev Mol Cell Biol. 2020. PMID: 32699358 No abstract available.
Similar articles
-
Dppa2 and Dppa4 counteract de novo methylation to establish a permissive epigenome for development.Nat Struct Mol Biol. 2020 Aug;27(8):706-716. doi: 10.1038/s41594-020-0445-1. Epub 2020 Jun 22. Nat Struct Mol Biol. 2020. PMID: 32572256
-
Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program.Genes Dev. 2019 Feb 1;33(3-4):194-208. doi: 10.1101/gad.321174.118. Epub 2019 Jan 28. Genes Dev. 2019. PMID: 30692203 Free PMC article.
-
Maternal Dppa2 and Dppa4 are dispensable for zygotic genome activation but important for offspring survival.Development. 2021 Dec 15;148(24):dev200191. doi: 10.1242/dev.200191. Epub 2021 Dec 21. Development. 2021. PMID: 34931676 Free PMC article.
-
Keeping your options open: insights from Dppa2/4 into how epigenetic priming factors promote cell plasticity.Biochem Soc Trans. 2020 Dec 18;48(6):2891-2902. doi: 10.1042/BST20200873. Biochem Soc Trans. 2020. PMID: 33336687 Free PMC article. Review.
-
Bivalent Histone Modifications and Development.Curr Stem Cell Res Ther. 2018;13(2):83-90. doi: 10.2174/1574888X12666170123144743. Curr Stem Cell Res Ther. 2018. PMID: 28117006 Review.
Cited by
-
DPPA2 and DPPA4 are dispensable for mouse zygotic genome activation and pre-implantation development.Development. 2021 Dec 15;148(24):dev200178. doi: 10.1242/dev.200178. Epub 2021 Dec 21. Development. 2021. PMID: 34878123 Free PMC article.
-
Loss of H3K9 trimethylation alters chromosome compaction and transcription factor retention during mitosis.Nat Struct Mol Biol. 2023 Apr;30(4):489-501. doi: 10.1038/s41594-023-00943-7. Epub 2023 Mar 20. Nat Struct Mol Biol. 2023. PMID: 36941433 Free PMC article.
-
Expanded Potential Stem Cells from Human Embryos Have an Open Chromatin Configuration with Enhanced Trophoblast Differentiation Ability.Adv Sci (Weinh). 2023 Apr;10(11):e2204797. doi: 10.1002/advs.202204797. Epub 2023 Feb 12. Adv Sci (Weinh). 2023. PMID: 36775869 Free PMC article.
-
A low-input high resolution sequential chromatin immunoprecipitation method captures genome-wide dynamics of bivalent chromatin.Epigenetics Chromatin. 2024 Feb 10;17(1):3. doi: 10.1186/s13072-024-00527-9. Epigenetics Chromatin. 2024. PMID: 38336688 Free PMC article.
-
Chromatin modifier developmental pluripotency associated factor 4 (DPPA4) is a candidate gene for alcohol-induced developmental disorders.BMC Med. 2022 Dec 30;20(1):495. doi: 10.1186/s12916-022-02699-1. BMC Med. 2022. PMID: 36581877 Free PMC article.
References
-
- Denissov S, et al. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Dev. 2014;141:526–537. - PubMed
-
- Bernstein BE, et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell. 2006;125:315–326. - PubMed
-
- Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

