Characterization of Extremely Drug-Resistant and Hypervirulent Acinetobacter baumannii AB030
- PMID: 32560407
- PMCID: PMC7345994
- DOI: 10.3390/antibiotics9060328
Characterization of Extremely Drug-Resistant and Hypervirulent Acinetobacter baumannii AB030
Abstract
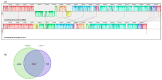
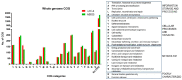
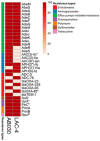
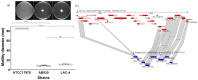
Acinetobacter baumannii is an important nosocomial bacterial pathogen. Multidrug-resistant isolates of A. baumannii are reported worldwide. Some A. baumannii isolates display resistance to nearly all antibiotics, making treatment of infections very challenging. As the need for new and effective antibiotics against A. baumannii becomes increasingly urgent, there is a need to understand the mechanisms of antibiotic resistance and virulence in this organism. In this work, comparative genomics was used to understand the mechanisms of antibiotic resistance and virulence in AB030, an extremely drug-resistant and hypervirulent strain of A. baumannii that is a representative of a recently emerged lineage of A. baumannii International Clone V. In order to characterize AB030, we carried out a genomic and phenotypic comparison with LAC-4, a previously described hyper-resistant and hypervirulent isolate. AB030 contains a number of antibiotic resistance- and virulence-associated genes that are not present in LAC-4. A number of these genes are present on mobile elements. This work shows the importance of characterizing the members of new lineages of A. baumannii in order to determine the development of antibiotic resistance and virulence in this organism.
Keywords: comparative genomics; gram-negative; insertion elements; multidrug resistance; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Complete genome sequence of hypervirulent and outbreak-associated Acinetobacter baumannii strain LAC-4: epidemiology, resistance genetic determinants and potential virulence factors.Sci Rep. 2015 Mar 2;5:8643. doi: 10.1038/srep08643. Sci Rep. 2015. PMID: 25728466 Free PMC article.
-
[Investigation of the virulence factors of multidrug-resistant Acinetobacter baumannii isolates].Mikrobiyol Bul. 2014 Jan;48(1):70-81. Mikrobiyol Bul. 2014. PMID: 24506717 Turkish.
-
Dissecting Colistin Resistance Mechanisms in Extensively Drug-Resistant Acinetobacter baumannii Clinical Isolates.mBio. 2019 Jul 16;10(4):e01083-19. doi: 10.1128/mBio.01083-19. mBio. 2019. PMID: 31311879 Free PMC article.
-
Interplay between Colistin Resistance, Virulence and Fitness in Acinetobacter baumannii.Antibiotics (Basel). 2017 Nov 21;6(4):28. doi: 10.3390/antibiotics6040028. Antibiotics (Basel). 2017. PMID: 29160808 Free PMC article. Review.
-
Insights into Acinetobacter baumannii: A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen.Antibiotics (Basel). 2020 Mar 12;9(3):119. doi: 10.3390/antibiotics9030119. Antibiotics (Basel). 2020. PMID: 32178356 Free PMC article. Review.
Cited by
-
The Acinetobacter baumannii website (Ab-web): a multidisciplinary knowledge hub, communication platform, and workspace.FEMS Microbes. 2023 Apr 21;4:xtad009. doi: 10.1093/femsmc/xtad009. eCollection 2023. FEMS Microbes. 2023. PMID: 37333444 Free PMC article.
-
Characterization of Virulent T4-Like Acinetobacter baumannii Bacteriophages DLP1 and DLP2.Viruses. 2023 Mar 13;15(3):739. doi: 10.3390/v15030739. Viruses. 2023. PMID: 36992448 Free PMC article.
-
Phylogenomic and phenotypic analyses highlight the diversity of antibiotic resistance and virulence in both human and non-human Acinetobacter baumannii.mSphere. 2024 Mar 26;9(3):e0074123. doi: 10.1128/msphere.00741-23. Epub 2024 Mar 5. mSphere. 2024. PMID: 38440986 Free PMC article.
-
Comparison of Hypervirulent and Non-Hypervirulent Carbapenem-Resistant Acinetobacter baumannii Isolated from Bloodstream Infections: Mortality, Potential Virulence Factors, and Combination Therapy In Vitro.Antibiotics (Basel). 2024 Aug 26;13(9):807. doi: 10.3390/antibiotics13090807. Antibiotics (Basel). 2024. PMID: 39334982 Free PMC article.
-
Characterization of a colistin resistant, hypervirulent hospital isolate of Acinetobacter courvalinii from Canada.Eur J Clin Microbiol Infect Dis. 2024 Oct;43(10):1939-1949. doi: 10.1007/s10096-024-04873-0. Epub 2024 Jul 29. Eur J Clin Microbiol Infect Dis. 2024. PMID: 39073669
References
-
- Chan P.C., Huang L.M., Lin H.C., Chang L.Y., Chen M.L., Lu C.Y., Lee P.I., Chen J.M., Lee C.Y., Pan H.J., et al. Control of an outbreak of pandrug-resistant Acinetobacter baumannii colonization and infection in a neonatal intensive care unit. Infect. Control Hosp. Epidemiol. 2007;28:423–429. doi: 10.1086/513120. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

