Common risk variants in NPHS1 and TNFSF15 are associated with childhood steroid-sensitive nephrotic syndrome
- PMID: 32554042
- PMCID: PMC8101291
- DOI: 10.1016/j.kint.2020.05.029
Common risk variants in NPHS1 and TNFSF15 are associated with childhood steroid-sensitive nephrotic syndrome
Abstract
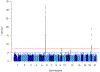
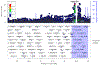
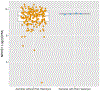
To understand the genetics of steroid-sensitive nephrotic syndrome (SSNS), we conducted a genome-wide association study in 987 childhood SSNS patients and 3,206 healthy controls with Japanese ancestry. Beyond known associations in the HLA-DR/DQ region, common variants in NPHS1-KIRREL2 (rs56117924, P=4.94E-20, odds ratio (OR) =1.90) and TNFSF15 (rs6478109, P=2.54E-8, OR=0.72) regions achieved genome-wide significance and were replicated in Korean, South Asian and African populations. Trans-ethnic meta-analyses including Japanese, Korean, South Asian, African, European, Hispanic and Maghrebian populations confirmed the significant associations of variants in NPHS1-KIRREL2 (Pmeta=6.71E-28, OR=1.88) and TNFSF15 (Pmeta=5.40E-11, OR=1.33) loci. Analysis of the NPHS1 risk alleles with glomerular NPHS1 mRNA expression from the same person revealed allele specific expression with significantly lower expression of the transcript derived from the risk haplotype (Wilcox test p=9.3E-4). Because rare pathogenic variants in NPHS1 cause congenital nephrotic syndrome of the Finnish type (CNSF), the present study provides further evidence that variation along the allele frequency spectrum in the same gene can cause or contribute to both a rare monogenic disease (CNSF) and a more complex, polygenic disease (SSNS).
Keywords: glomerulus; nephrotic syndrome; pediatric nephrology; podocyte.
Copyright © 2020 International Society of Nephrology. Published by Elsevier Inc. All rights reserved.
Figures





Similar articles
-
An updated view of the pathogenesis of steroid-sensitive nephrotic syndrome.Pediatr Nephrol. 2022 Sep;37(9):1957-1965. doi: 10.1007/s00467-021-05401-4. Epub 2022 Jan 10. Pediatr Nephrol. 2022. PMID: 35006356 Free PMC article. Review.
-
Strong Association of the HLA-DR/DQ Locus with Childhood Steroid-Sensitive Nephrotic Syndrome in the Japanese Population.J Am Soc Nephrol. 2018 Aug;29(8):2189-2199. doi: 10.1681/ASN.2017080859. Epub 2018 Jul 16. J Am Soc Nephrol. 2018. PMID: 30012571 Free PMC article.
-
HLA-DQA1 and APOL1 as Risk Loci for Childhood-Onset Steroid-Sensitive and Steroid-Resistant Nephrotic Syndrome.Am J Kidney Dis. 2018 Mar;71(3):399-406. doi: 10.1053/j.ajkd.2017.10.013. Epub 2017 Dec 23. Am J Kidney Dis. 2018. PMID: 29277510 Free PMC article.
-
HLA-DQA1 and PLCG2 Are Candidate Risk Loci for Childhood-Onset Steroid-Sensitive Nephrotic Syndrome.J Am Soc Nephrol. 2015 Jul;26(7):1701-10. doi: 10.1681/ASN.2014030247. Epub 2014 Oct 27. J Am Soc Nephrol. 2015. PMID: 25349203 Free PMC article.
-
Novel insights in the genetics of steroid-sensitive nephrotic syndrome in childhood.Pediatr Nephrol. 2021 Aug;36(8):2165-2175. doi: 10.1007/s00467-020-04780-4. Epub 2020 Oct 21. Pediatr Nephrol. 2021. PMID: 33084934 Review.
Cited by
-
Quantifiable and reproducible phenotypic assessment of a constitutive knockout mouse model for congenital nephrotic syndrome of the Finnish type.Sci Rep. 2024 Jul 10;14(1):15916. doi: 10.1038/s41598-024-64883-y. Sci Rep. 2024. PMID: 38987283 Free PMC article.
-
Common Risk Variants in AHI1 Are Associated With Childhood Steroid Sensitive Nephrotic Syndrome.Kidney Int Rep. 2023 May 27;8(8):1562-1574. doi: 10.1016/j.ekir.2023.05.018. eCollection 2023 Aug. Kidney Int Rep. 2023. PMID: 37547536 Free PMC article.
-
B-Cell Dysregulation in Idiopathic Nephrotic Syndrome: What We Know and What We Need to Discover.Front Immunol. 2022 Jan 24;13:823204. doi: 10.3389/fimmu.2022.823204. eCollection 2022. Front Immunol. 2022. PMID: 35140723 Free PMC article. Review.
-
Glomerular Diseases of the Kidney Allograft: Toward a Precision Medicine Approach.Semin Nephrol. 2022 Jan;42(1):29-43. doi: 10.1016/j.semnephrol.2022.01.005. Semin Nephrol. 2022. PMID: 35618394 Free PMC article. Review.
-
T-cell receptor diversity in minimal change disease in the NEPTUNE study.Pediatr Nephrol. 2023 Apr;38(4):1115-1126. doi: 10.1007/s00467-022-05696-x. Epub 2022 Aug 9. Pediatr Nephrol. 2023. PMID: 35943576 Free PMC article.
References
-
- White RH The familial nephrotic syndrome. I. A European survey. Clin Nephrol. 1973;1:215–219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

