The Transcription Co-Repressors MTG8 and MTG16 Regulate Exit of Intestinal Stem Cells From Their Niche and Differentiation Into Enterocyte vs Secretory Lineages
- PMID: 32553763
- PMCID: PMC7607384
- DOI: 10.1053/j.gastro.2020.06.012
The Transcription Co-Repressors MTG8 and MTG16 Regulate Exit of Intestinal Stem Cells From Their Niche and Differentiation Into Enterocyte vs Secretory Lineages
Abstract
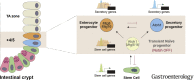
Background & aims: Notch signaling maintains intestinal stem cells (ISCs). When ISCs exit the niche, Notch signaling among early progenitor cells at position +4/5 regulates their specification toward secretory vs enterocyte lineages (binary fate). The transcription factor ATOH1 is repressed by Notch in ISCs; its de-repression, when Notch is inactivated, drives progenitor cells to differentiate along the secretory lineage. However, it is not clear what promotes transition of ISCs to progenitors and how this fate decision is established.
Methods: We sorted cells from Lgr5-GFP knockin intestines from mice and characterized gene expression patterns. We analyzed Notch regulation by examining expression profiles (by quantitative reverse transcription polymerase chain reaction and RNAscope) of small intestinal organoids incubated with the Notch inhibitor DAPT, intestine tissues from mice given injections of the γ-secretase inhibitor dibenzazepine, and mice with intestine-specific disruption of Rbpj. We analyzed intestine tissues from mice with disruption of the RUNX1 translocation partner 1 gene (Runx1t1, also called Mtg8) or CBFA2/RUNX1 partner transcriptional co-repressor 3 (Cbfa2t3, also called Mtg16), and derived their organoids, by histology, immunohistochemistry, and RNA sequencing (RNA-seq). We performed chromatin immunoprecipitation and sequencing analyses of intestinal crypts to identify genes regulated by MTG16.
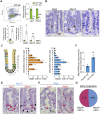
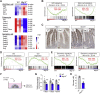
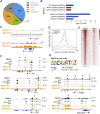
Results: The transcription co-repressors MTG8 and MTG16 were highly expressed by +4/5 early progenitors, compared with other cells along crypt-villus axis. Expression of MTG8 and MTG16 were repressed by Notch signaling via ATOH1 in organoids and intestine tissues from mice. MTG8- and MTG16-knockout intestines had increased crypt hyperproliferation and expansion of ISCs, but enterocyte differentiation was impaired, based on loss of enterocyte markers and functions. Chromatin immunoprecipitation and sequencing analyses showed that MTG16 bound to promoters of genes that are specifically expressed by stem cells (such as Lgr5 and Ascl2) and repressed their transcription. MTG16 also bound to previously reported enhancer regions of genes regulated by ATOH1, including genes that encode Delta-like canonical Notch ligand and other secretory-specific transcription factors.
Conclusions: In intestine tissues of mice and human intestinal organoids, MTG8 and MTG16 repress transcription in the earliest progenitor cells to promote exit of ISCs from their niche (niche exit) and control the binary fate decision (secretory vs enterocyte lineage) by repressing genes regulated by ATOH1.
Keywords: Chromatin Remodeling; Lateral Inhibition; Lineage Specification; Niche Exit.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
LKB1 Represses ATOH1 via PDK4 and Energy Metabolism and Regulates Intestinal Stem Cell Fate.Gastroenterology. 2020 Apr;158(5):1389-1401.e10. doi: 10.1053/j.gastro.2019.12.033. Epub 2020 Jan 11. Gastroenterology. 2020. PMID: 31930988
-
BHLHA15-Positive Secretory Precursor Cells Can Give Rise to Tumors in Intestine and Colon in Mice.Gastroenterology. 2019 Mar;156(4):1066-1081.e16. doi: 10.1053/j.gastro.2018.11.024. Epub 2018 Nov 15. Gastroenterology. 2019. PMID: 30448068 Free PMC article.
-
Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity.Nature. 2014 Feb 27;506(7489):511-5. doi: 10.1038/nature12903. Epub 2014 Jan 12. Nature. 2014. PMID: 24413398 Free PMC article.
-
Notch Signaling in Mammalian Intestinal Stem Cells: Determining Cell Fate and Maintaining Homeostasis.Curr Stem Cell Res Ther. 2019;14(7):583-590. doi: 10.2174/1574888X14666190429143734. Curr Stem Cell Res Ther. 2019. PMID: 31729290 Review.
-
Signaling and epigenetic mechanisms of intestinal stem cells and progenitors: insight into crypt homeostasis, plasticity, and niches.Wiley Interdiscip Rev Dev Biol. 2017 Sep;6(5). doi: 10.1002/wdev.281. Epub 2017 Jun 23. Wiley Interdiscip Rev Dev Biol. 2017. PMID: 28644919 Review.
Cited by
-
Control of Paneth cell function by HuR regulates gut mucosal growth by altering stem cell activity.Life Sci Alliance. 2023 Sep 11;6(11):e202302152. doi: 10.26508/lsa.202302152. Print 2023 Nov. Life Sci Alliance. 2023. PMID: 37696579 Free PMC article.
-
Loss of ARID3A perturbs intestinal epithelial proliferation-differentiation ratio and regeneration.J Exp Med. 2024 Oct 7;221(10):e20232279. doi: 10.1084/jem.20232279. Epub 2024 Aug 16. J Exp Med. 2024. PMID: 39150450 Free PMC article.
-
MTG16 regulates colonic epithelial differentiation, colitis, and tumorigenesis by repressing E protein transcription factors.JCI Insight. 2022 May 23;7(10):e153045. doi: 10.1172/jci.insight.153045. JCI Insight. 2022. PMID: 35503250 Free PMC article.
-
RUNX1T1 function in cell fate.Stem Cell Res Ther. 2022 Jul 28;13(1):369. doi: 10.1186/s13287-022-03074-w. Stem Cell Res Ther. 2022. PMID: 35902872 Free PMC article. Review.
-
Advanced Progression for the Heterogeneity and Homeostasis of Intestinal Stem Cells.Stem Cell Rev Rep. 2023 Oct;19(7):2109-2119. doi: 10.1007/s12015-023-10578-2. Epub 2023 Jun 23. Stem Cell Rev Rep. 2023. PMID: 37351833 Review.
References
-
- Bjerknes M., Cheng H. The stem-cell zone of the small intestinal epithelium. V. Evidence for controls over orientation of boundaries between the stem-cell zone, proliferative zone, and the maturation zone. Am J Anat. 1981;160:105–112. - PubMed
-
- Tetteh P.W., Farin H.F., Clevers H. Plasticity within stem cell hierarchies in mammalian epithelia. Trends Cell Biol. 2015;25:100–108. - PubMed
-
- Yang Q., Bermingham N.A., Finegold M.J. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. - PubMed
-
- van Es J.H., van Gijn M.E., Riccio O. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. - PubMed
Supplementary References
-
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 2011;17:3.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

