Structural and Functional Annotation of Transposable Elements Revealed a Potential Regulation of Genes Involved in Rubber Biosynthesis by TE-Derived siRNA Interference in Hevea brasiliensis
- PMID: 32545790
- PMCID: PMC7353026
- DOI: 10.3390/ijms21124220
Structural and Functional Annotation of Transposable Elements Revealed a Potential Regulation of Genes Involved in Rubber Biosynthesis by TE-Derived siRNA Interference in Hevea brasiliensis
Abstract
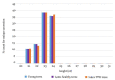
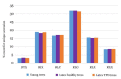
The natural rubber biosynthetic pathway is well described in Hevea, although the final stages of rubber elongation are still poorly understood. Small Rubber Particle Proteins and Rubber Elongation Factors (SRPPs and REFs) are proteins with major function in rubber particle formation and stabilization. Their corresponding genes are clustered on a scaffold1222 of the reference genomic sequence of the Hevea brasiliensis genome. Apart from gene expression by transcriptomic analyses, to date, no deep analyses have been carried out for the genomic environment of SRPPs and REFs loci. By integrative analyses on transposable element annotation, small RNAs production and gene expression, we analysed their role in the control of the transcription of rubber biosynthetic genes. The first in-depth annotation of TEs (Transposable Elements) and their capacity to produce TE-derived siRNAs (small interfering RNAs) is presented, only possible in the Hevea brasiliensis clone PB 260 for which all data are available. We observed that 11% of genes are located near TEs and their presence may interfere in their transcription at both genetic and epigenetic level. We hypothesized that the genomic environment of rubber biosynthesis genes has been shaped by TE and TE-derived siRNAs with possible transcriptional interference on their gene expression. We discussed possible functionalization of TEs as enhancers and as donors of alternative transcription start sites in promoter sequences, possibly through the modelling of genetic and epigenetic landscapes.
Keywords: epigenomics; rubber tree; siRNA; transcriptional regulation; transposable elements.
Conflict of interest statement
The authors declare no conflict of interest and the funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


Similar articles
-
Large-scale collection of full-length cDNA and transcriptome analysis in Hevea brasiliensis.DNA Res. 2017 Apr 1;24(2):159-167. doi: 10.1093/dnares/dsw056. DNA Res. 2017. PMID: 28431015 Free PMC article.
-
Transcriptome analyses reveal molecular mechanism underlying tapping panel dryness of rubber tree (Hevea brasiliensis).Sci Rep. 2016 Mar 23;6:23540. doi: 10.1038/srep23540. Sci Rep. 2016. PMID: 27005401 Free PMC article.
-
Impact of an intragenic retrotransposon on the structural integrity and evolution of a major isoprenoid biosynthesis pathway gene in Hevea brasiliensis.Plant Physiol Biochem. 2013 Dec;73:176-88. doi: 10.1016/j.plaphy.2013.09.004. Epub 2013 Sep 24. Plant Physiol Biochem. 2013. PMID: 24128694
-
Genomic technologies for Hevea breeding.Adv Genet. 2019;104:1-73. doi: 10.1016/bs.adgen.2019.04.001. Epub 2019 Jun 3. Adv Genet. 2019. PMID: 31200808 Review.
-
Hevea brasiliensis REF (Hev b 1) and SRPP (Hev b 3): An overview on rubber particle proteins.Biochimie. 2014 Nov;106:1-9. doi: 10.1016/j.biochi.2014.07.002. Epub 2014 Jul 11. Biochimie. 2014. PMID: 25019490 Review.
Cited by
-
The rubber tree kinome: Genome-wide characterization and insights into coexpression patterns associated with abiotic stress responses.Front Plant Sci. 2023 Feb 7;14:1068202. doi: 10.3389/fpls.2023.1068202. eCollection 2023. Front Plant Sci. 2023. PMID: 36824205 Free PMC article.
-
Reviving Natural Rubber Synthesis via Native/Large Nanodiscs.Polymers (Basel). 2024 May 22;16(11):1468. doi: 10.3390/polym16111468. Polymers (Basel). 2024. PMID: 38891415 Free PMC article. Review.
-
Unravelling Rubber Tree Growth by Integrating GWAS and Biological Network-Based Approaches.Front Plant Sci. 2021 Dec 21;12:768589. doi: 10.3389/fpls.2021.768589. eCollection 2021. Front Plant Sci. 2021. PMID: 34992619 Free PMC article.
References
-
- Compagnon P., Chapuset T., Gener P., Jacob J.-L., De La Serve M., De Livonnière H., Nicolas D., Omont H., Serier J.-B., Tran Van Canh C., et al. Le Caoutchouc Naturel Biologie, Culture, Production. Maisonneuve et Larose; Paris, France: 1986. p. 595.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

