The Contorsbody, an antibody format for agonism: Design, structure, and function
- PMID: 32542107
- PMCID: PMC7283085
- DOI: 10.1016/j.csbj.2020.05.007
The Contorsbody, an antibody format for agonism: Design, structure, and function
Abstract
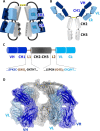
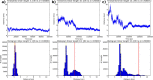
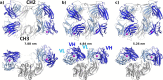
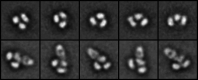
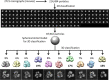
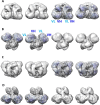
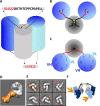
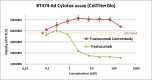
The careful design of the antibody architecture is becoming more and more important, especially when the purpose is agonism. We present the design of a novel antibody format that is able to promote receptor dimerization and induce signal transduction resulting in cell proliferation. Mono-specific bivalent Y-shape IgGs made of two light chains and two heavy chains are engineered into single chain dimers of two modified heavy chains, resulting in the fixation of the two Fab fragments along the Fc dimerizing moiety. By this, an antagonist of the Her-receptor family, Trastuzumab, is re-formatted into an agonist by simply incorporating the original binding motif into a different geometrically and sterically constrained conformation. This novel format, named Contorsbody, retains antigen binding properties of the parental IgGs and can be produced by standard technologies established for recombinant IgGs. Structural analyses using molecular dynamics and electron microscopy are described to guide the initial design and to confirm the Contorsbody as a very compact molecule, respectively. Contorsbodies show increased rigidity compared to IgGs and their Fab moieties are positioned parallel and adjacent to each other. This geometry has an increased potential to trigger cell surface antigen or receptor 'cis'-dimerization without 'trans'-bridging of cells or mere receptor blockade.
Keywords: Antibody format; Electron microscopy; Molecular dynamic; Molecular modeling; Protein engineering; Receptor dimerization.
© 2020 The Author(s).
Figures


Similar articles
-
Format and geometries matter: Structure-based design defines the functionality of bispecific antibodies.Comput Struct Biotechnol J. 2020 May 14;18:1221-1227. doi: 10.1016/j.csbj.2020.05.006. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32542108 Free PMC article. Review.
-
"BIClonals": Production of Bispecific Antibodies in IgG Format in Transiently Transfected Mammalian Cells.Methods Mol Biol. 2019;1904:431-454. doi: 10.1007/978-1-4939-8958-4_22. Methods Mol Biol. 2019. PMID: 30539485
-
Spatial structure of immunoglobulin molecules.Klin Wochenschr. 1980 Nov 17;58(22):1217-31. doi: 10.1007/BF01478928. Klin Wochenschr. 1980. PMID: 6780722 Review.
-
Bivalent antibody phage display mimics natural immunoglobulin.J Immunol Methods. 2004 Jan;284(1-2):119-32. doi: 10.1016/j.jim.2003.11.001. J Immunol Methods. 2004. PMID: 14736422
-
An improved single-chain Fab platform for efficient display and recombinant expression.J Mol Biol. 2015 Jan 30;427(2):576-86. doi: 10.1016/j.jmb.2014.11.017. Epub 2014 Dec 3. J Mol Biol. 2015. PMID: 25481745 Free PMC article.
Cited by
-
Binding domain on CD22 molecules contributing to the biological activity of T cell-engaging bispecific antibodies.Heliyon. 2023 Jul 4;9(7):e17960. doi: 10.1016/j.heliyon.2023.e17960. eCollection 2023 Jul. Heliyon. 2023. PMID: 37456045 Free PMC article.
-
Format and geometries matter: Structure-based design defines the functionality of bispecific antibodies.Comput Struct Biotechnol J. 2020 May 14;18:1221-1227. doi: 10.1016/j.csbj.2020.05.006. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32542108 Free PMC article. Review.
-
Radiolabelling small and biomolecules for tracking and monitoring.RSC Adv. 2022 Nov 11;12(50):32383-32400. doi: 10.1039/d2ra06236d. eCollection 2022 Nov 9. RSC Adv. 2022. PMID: 36425706 Free PMC article. Review.
-
TFAB002s, novel CD20-targeting T cell-dependent bispecific Fab-FabCH3 antibodies, exhibit potent antitumor efficacy against malignant B-cell lymphoma.PLoS One. 2024 Sep 25;19(9):e0310889. doi: 10.1371/journal.pone.0310889. eCollection 2024. PLoS One. 2024. PMID: 39321199 Free PMC article.
-
Format chain exchange (FORCE) for high-throughput generation of bispecific antibodies in combinatorial binder-format matrices.Nat Commun. 2020 Oct 2;11(1):4974. doi: 10.1038/s41467-020-18477-7. Nat Commun. 2020. PMID: 33009381 Free PMC article.
References
-
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. doi: 10.1016/j.softx.2015.06.001. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
