Location, Location, Location: The Role of Nuclear Positioning in the Repair of Collapsed Forks and Protection of Genome Stability
- PMID: 32526925
- PMCID: PMC7348918
- DOI: 10.3390/genes11060635
Location, Location, Location: The Role of Nuclear Positioning in the Repair of Collapsed Forks and Protection of Genome Stability
Abstract
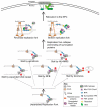
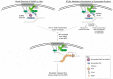
Components of the nuclear pore complex (NPC) have been shown to play a crucial role in protecting against replication stress, and recovery from some types of stalled or collapsed replication forks requires movement of the DNA to the NPC in order to maintain genome stability. The role that nuclear positioning has on DNA repair has been investigated in several systems that inhibit normal replication. These include structure forming sequences (expanded CAG repeats), protein mediated stalls (replication fork barriers (RFBs)), stalls within the telomere sequence, and the use of drugs known to stall or collapse replication forks (HU + MMS or aphidicolin). Recently, the mechanism of relocation for collapsed replication forks to the NPC has been elucidated. Here, we will review the types of replication stress that relocate to the NPC, the current models for the mechanism of relocation, and the currently known protective effects of this movement.
Keywords: fork collapse; fork restart; nuclear pore complex; replication fork; replication fork barriers; sumoylation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Relocation of Collapsed Forks to the Nuclear Pore Complex Depends on Sumoylation of DNA Repair Proteins and Permits Rad51 Association.Cell Rep. 2020 May 12;31(6):107635. doi: 10.1016/j.celrep.2020.107635. Cell Rep. 2020. PMID: 32402281 Free PMC article.
-
Relocalization of DNA lesions to the nuclear pore complex.FEMS Yeast Res. 2016 Dec 1;16(8):fow095. doi: 10.1093/femsyr/fow095. FEMS Yeast Res. 2016. PMID: 27799300 Free PMC article. Review.
-
A tough row to hoe: when replication forks encounter DNA damage.Biochem Soc Trans. 2018 Dec 17;46(6):1643-1651. doi: 10.1042/BST20180308. Epub 2018 Dec 4. Biochem Soc Trans. 2018. PMID: 30514768 Free PMC article. Review.
-
The DNA Replication Checkpoint Targets the Kinetochore for Relocation of Collapsed Forks to the Nuclear Periphery.bioRxiv [Preprint]. 2024 Jul 4:2024.06.17.599319. doi: 10.1101/2024.06.17.599319. bioRxiv. 2024. PMID: 38948692 Free PMC article. Preprint.
-
Restarted replication forks are error-prone and cause CAG repeat expansions and contractions.PLoS Genet. 2021 Oct 21;17(10):e1009863. doi: 10.1371/journal.pgen.1009863. eCollection 2021 Oct. PLoS Genet. 2021. PMID: 34673780 Free PMC article.
Cited by
-
Telomere Replication: Solving Multiple End Replication Problems.Front Cell Dev Biol. 2021 Apr 1;9:668171. doi: 10.3389/fcell.2021.668171. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33869233 Free PMC article. Review.
-
MT1-MMP-dependent ECM processing regulates laminB1 stability and mediates replication fork restart.PLoS One. 2021 Jul 8;16(7):e0253062. doi: 10.1371/journal.pone.0253062. eCollection 2021. PLoS One. 2021. PMID: 34237080 Free PMC article.
-
STIM1 translocation to the nucleus protects cells from DNA damage.Nucleic Acids Res. 2024 Mar 21;52(5):2389-2415. doi: 10.1093/nar/gkae001. Nucleic Acids Res. 2024. PMID: 38224453 Free PMC article.
-
Determinants of RPA megafoci localization to the nuclear periphery in response to replication stress.G3 (Bethesda). 2022 Jul 6;12(7):jkac116. doi: 10.1093/g3journal/jkac116. G3 (Bethesda). 2022. PMID: 35567482 Free PMC article.
-
Nuclear Lipid Droplet Birth during Replicative Stress.Cells. 2022 Apr 20;11(9):1390. doi: 10.3390/cells11091390. Cells. 2022. PMID: 35563696 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

