Circ_0000517 Contributes to Hepatocellular Carcinoma Progression by Upregulating TXNDC5 via Sponging miR-1296-5p
- PMID: 32523376
- PMCID: PMC7234960
- DOI: 10.2147/CMAR.S244024
Circ_0000517 Contributes to Hepatocellular Carcinoma Progression by Upregulating TXNDC5 via Sponging miR-1296-5p
Abstract
Background: Circular RNAs (circRNAs) function as essential regulators in diverse human cancers, including hepatocellular carcinoma (HCC). However, the function of circ_0000517 in HCC was unknown. We aimed to explore the roles and mechanisms of circ_0000517 in HCC.
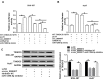
Materials and methods: The levels of circ_0000517, RPPH1 mRNA and microRNA-1296-5p (miR-1296-5p) were measured using quantitative real-time polymerase chain reaction (qRT-PCR). The characteristics of circ_0000517 were explored by RNase R digestion and actinomycin D assays. Cell proliferation was evaluated by Cell Counting Kit-8 (CCK-8) and colony formation assays. Cell cycle process and cell apoptosis were analyzed by flow cytometry analysis. The function of circ_0000517 in vivo was explored by a murine xenograft model. The association between miR-1296-5p and circ_0000517 or thioredoxin domain containing 5 (TXNDC5) was determined by dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay. The protein level of TXNDC5 was detected by Western blot assay.
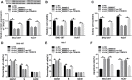
Results: Circ_0000517 was upregulated in HCC tissues and cells. Silencing of circ_0000517 suppressed HCC cell viability and colony formation and promoted cell cycle arrest and apoptosis in vitro and hampered tumor growth in vivo. MiR-1296-5p was a target of circ_0000517 and the effects of circ_0000517 silencing on HCC cell viability, cell cycle, colony formation and apoptosis were abolished by miR-1296-5p inhibition. TXNDC5 functioned as a target gene of miR-1296-5p, and the inhibitory effect of miR-1296-5p on HCC cell progression was rescued by TXNDC5 overexpression. Moreover, circ_0000517 promoted TXNDC5 expression via targeting miR-1296-5p.
Conclusion: Circ_0000517 accelerated HCC progression by upregulating TXNDC5 through sponging miR-1296-5p.
Keywords: HCC; TXNDC5; circ_0000517; miR-1296-5p.
© 2020 Zang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest in this work.
Figures




Similar articles
-
Circ_0078710 promotes the development of liver cancer by upregulating TXNDC5 via miR-431-5p.Ann Hepatol. 2022 Jan-Feb;27(1):100551. doi: 10.1016/j.aohep.2021.100551. Epub 2021 Oct 2. Ann Hepatol. 2022. PMID: 34606982
-
Circ_0011232 contributes to hepatocellular carcinoma progression through miR-503-5p/AKT3 axis.Hepatol Res. 2022 Jun;52(6):532-545. doi: 10.1111/hepr.13758. Epub 2022 Mar 28. Hepatol Res. 2022. PMID: 35187761
-
Circ_0074027 Contributes to Non-Small Cell Lung Cancer Progression by Upregulating CUL4B Expression Through miR-335-5p.Cancer Biother Radiopharm. 2022 Mar;37(2):73-83. doi: 10.1089/cbr.2020.3579. Epub 2020 Jun 22. Cancer Biother Radiopharm. 2022. PMID: 32580576
-
Circ-CSPP1 knockdown suppresses hepatocellular carcinoma progression through miR-493-5p releasing-mediated HMGB1 downregulation.Cell Signal. 2021 Oct;86:110065. doi: 10.1016/j.cellsig.2021.110065. Epub 2021 Jun 26. Cell Signal. 2021. PMID: 34182091 Review.
-
Thioredoxin Domain Containing 5 (TXNDC5): Friend or Foe?Curr Issues Mol Biol. 2024 Apr 4;46(4):3134-3163. doi: 10.3390/cimb46040197. Curr Issues Mol Biol. 2024. PMID: 38666927 Free PMC article. Review.
Cited by
-
Protein Disulfide Isomerases Function as the Missing Link Between Diabetes and Cancer.Antioxid Redox Signal. 2022 Dec;37(16-18):1191-1205. doi: 10.1089/ars.2022.0098. Epub 2022 Nov 21. Antioxid Redox Signal. 2022. PMID: 36000195 Free PMC article. Review.
-
Circular RNA circ_0000517 Facilitates The Growth and Metastasis of Non-Small Cell Lung Cancer by Sponging miR-326/miR-330-5p.Cell J. 2021 Oct;23(5):552-561. doi: 10.22074/cellj.2021.7913. Epub 2021 Oct 30. Cell J. 2021. PMID: 34837683 Free PMC article.
-
Silencing of circRPPH1 Inhibits the Progression of Non-Small-Cell Lung Cancer Through miR-326/ERBB4 Signal Axis.Biochem Genet. 2024 May 22. doi: 10.1007/s10528-024-10824-3. Online ahead of print. Biochem Genet. 2024. PMID: 38776052
-
Thioredoxin Domain Containing 5 Suppression Elicits Serum Amyloid A-Containing High-Density Lipoproteins.Biomedicines. 2022 Mar 18;10(3):709. doi: 10.3390/biomedicines10030709. Biomedicines. 2022. PMID: 35327511 Free PMC article.
-
Hsa_circ_0017639 regulates cisplatin resistance and tumor growth via acting as a miR-1296-5p molecular sponge and modulating sine oculis homeobox 1 expression in non-small cell lung cancer.Bioengineered. 2022 Apr;13(4):8806-8822. doi: 10.1080/21655979.2022.2053810. Bioengineered. 2022. PMID: 35287543 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

