A cancer drug atlas enables synergistic targeting of independent drug vulnerabilities
- PMID: 32523045
- PMCID: PMC7287046
- DOI: 10.1038/s41467-020-16735-2
A cancer drug atlas enables synergistic targeting of independent drug vulnerabilities
Abstract
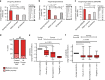
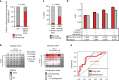
Personalized cancer treatments using combinations of drugs with a synergistic effect is attractive but proves to be highly challenging. Here we present an approach to uncover the efficacy of drug combinations based on the analysis of mono-drug effects. For this we used dose-response data from pharmacogenomic encyclopedias and represent these as a drug atlas. The drug atlas represents the relations between drug effects and allows to identify independent processes for which the tumor might be particularly vulnerable when attacked by two drugs. Our approach enables the prediction of combination-therapy which can be linked to tumor-driving mutations. By using this strategy, we can uncover potential effective drug combinations on a pan-cancer scale. Predicted synergies are provided and have been validated in glioblastoma, breast cancer, melanoma and leukemia mouse-models, resulting in therapeutic synergy in 75% of the tested models. This indicates that we can accurately predict effective drug combinations with translational value.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Combination drug screen targeting glioblastoma core vulnerabilities reveals pharmacological synergisms.EBioMedicine. 2023 Sep;95:104752. doi: 10.1016/j.ebiom.2023.104752. Epub 2023 Aug 10. EBioMedicine. 2023. PMID: 37572644 Free PMC article.
-
An integrated framework for identification of effective and synergistic anti-cancer drug combinations.J Bioinform Comput Biol. 2018 Oct;16(5):1850017. doi: 10.1142/S0219720018500178. Epub 2018 Jun 28. J Bioinform Comput Biol. 2018. PMID: 30304987
-
Patient-Customized Drug Combination Prediction and Testing for T-cell Prolymphocytic Leukemia Patients.Cancer Res. 2018 May 1;78(9):2407-2418. doi: 10.1158/0008-5472.CAN-17-3644. Epub 2018 Feb 26. Cancer Res. 2018. PMID: 29483097
-
Stratification and prediction of drug synergy based on target functional similarity.NPJ Syst Biol Appl. 2020 Jun 2;6(1):16. doi: 10.1038/s41540-020-0136-x. NPJ Syst Biol Appl. 2020. PMID: 32487991 Free PMC article.
-
A review of machine learning approaches for drug synergy prediction in cancer.Brief Bioinform. 2022 May 13;23(3):bbac075. doi: 10.1093/bib/bbac075. Brief Bioinform. 2022. PMID: 35323854 Review.
Cited by
-
Bipartite network models to design combination therapies in acute myeloid leukaemia.Nat Commun. 2022 Apr 19;13(1):2128. doi: 10.1038/s41467-022-29793-5. Nat Commun. 2022. PMID: 35440130 Free PMC article.
-
Hybrid peptides as platform for synchronized combination therapy.Colloids Surf B Biointerfaces. 2023 Jun;226:113326. doi: 10.1016/j.colsurfb.2023.113326. Epub 2023 Apr 24. Colloids Surf B Biointerfaces. 2023. PMID: 37116378 Free PMC article.
-
CombPDX: a unified statistical framework for evaluating drug synergism in patient-derived xenografts.Sci Rep. 2022 Jul 29;12(1):12984. doi: 10.1038/s41598-022-16933-6. Sci Rep. 2022. PMID: 35906256 Free PMC article.
-
Terpenes-Modified Lipid Nanosystems for Temozolomide, Improving Cytotoxicity against Glioblastoma Human Cancer Cells In Vitro.Nanomaterials (Basel). 2023 Dec 24;14(1):55. doi: 10.3390/nano14010055. Nanomaterials (Basel). 2023. PMID: 38202510 Free PMC article.
-
A technical note on emerging combination approach involved in the onconanotherapeutics.Drug Deliv. 2022 Dec;29(1):3197-3212. doi: 10.1080/10717544.2022.2132018. Drug Deliv. 2022. PMID: 36226570 Free PMC article. Review.
References
-
- Hatzivassiliou G, et al. ERK inhibition overcomes acquired resistance to MEK inhibitors. Mol. Cancer Ther. 2012;11:1143–1154. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

