This is a preprint.
Predicting the Immunogenicity of T cell epitopes: From HIV to SARS-CoV-2
- PMID: 32511339
- PMCID: PMC7241102
- DOI: 10.1101/2020.05.14.095885
Predicting the Immunogenicity of T cell epitopes: From HIV to SARS-CoV-2
Update in
-
Learning from HIV-1 to predict the immunogenicity of T cell epitopes in SARS-CoV-2.iScience. 2021 Apr 23;24(4):102311. doi: 10.1016/j.isci.2021.102311. Epub 2021 Mar 15. iScience. 2021. PMID: 33748696 Free PMC article.
Abstract
We describe a physics-based learning model for predicting the immunogenicity of Cytotoxic T Lymphocyte (CTL) epitopes derived from diverse pathogens, given a Human Leukocyte Antigen (HLA) genotype. The model was trained and tested on experimental data on the relative immunodominance of CTL epitopes in Human Immunodeficiency Virus infection. The method is more accurate than publicly available models. Our model predicts that only a fraction of SARS-CoV-2 epitopes that have been predicted to bind to HLA molecules is immunogenic. The immunogenic CTL epitopes across all SARS-CoV-2 proteins are predicted to provide broad population coverage, but the immunogenic epitopes in the SARS-CoV-2 spike protein alone are unlikely to do so. Our model predicts that several immunogenic SARS-CoV-2 CTL epitopes are identical to those contained in low-pathogenicity coronaviruses circulating in the population. Thus, we suggest that some level of CTL immunity against COVID-19 may be present in some individuals prior to SARS-CoV-2 infection.
Figures
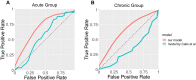
Similar articles
-
Learning from HIV-1 to predict the immunogenicity of T cell epitopes in SARS-CoV-2.iScience. 2021 Apr 23;24(4):102311. doi: 10.1016/j.isci.2021.102311. Epub 2021 Mar 15. iScience. 2021. PMID: 33748696 Free PMC article.
-
COVID-19 coronavirus vaccine T cell epitope prediction analysis based on distributions of HLA class I loci (HLA-A, -B, -C) across global populations.Hum Vaccin Immunother. 2021 Apr 3;17(4):1097-1108. doi: 10.1080/21645515.2020.1823777. Epub 2020 Nov 11. Hum Vaccin Immunother. 2021. PMID: 33175614 Free PMC article.
-
Sequence-based prediction of SARS-CoV-2 vaccine targets using a mass spectrometry-based bioinformatics predictor identifies immunogenic T cell epitopes.Genome Med. 2020 Aug 13;12(1):70. doi: 10.1186/s13073-020-00767-w. Genome Med. 2020. PMID: 32791978 Free PMC article.
-
Evolution of SARS-CoV-2-specific CD4+ T cell epitopes.Immunogenetics. 2023 Jun;75(3):283-293. doi: 10.1007/s00251-023-01295-8. Epub 2023 Jan 31. Immunogenetics. 2023. PMID: 36719467 Free PMC article. Review.
-
Identification of Novel Candidate Epitopes on SARS-CoV-2 Proteins for South America: A Review of HLA Frequencies by Country.Front Immunol. 2020 Sep 3;11:2008. doi: 10.3389/fimmu.2020.02008. eCollection 2020. Front Immunol. 2020. PMID: 33013857 Free PMC article. Review.
Cited by
-
Identification of immunodominant linear epitopes from SARS-CoV-2 patient plasma.PLoS One. 2020 Sep 9;15(9):e0238089. doi: 10.1371/journal.pone.0238089. eCollection 2020. PLoS One. 2020. PMID: 32903266 Free PMC article.
References
-
- Akst J. (2020) ‘COVID-19 Vaccine Frontrunners’, TheScientist. doi: https://www.the-scientist.com/news-opinion/covid-19-vaccine-frontrunners....
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
