HIV-1 induced changes in HLA-C*03 : 04-presented peptide repertoires lead to reduced engagement of inhibitory natural killer cell receptors
- PMID: 32501836
- PMCID: PMC8635260
- DOI: 10.1097/QAD.0000000000002596
HIV-1 induced changes in HLA-C*03 : 04-presented peptide repertoires lead to reduced engagement of inhibitory natural killer cell receptors
Abstract
Objective: Viral infections influence intracellular peptide repertoires available for presentation by HLA-I. Alterations in HLA-I/peptide complexes can modulate binding of killer immunoglobuline-like receptors (KIRs) and thereby the function of natural killer (NK) cells. Although multiple studies have provided evidence that HLA-I/KIR interactions play a role in HIV-1 disease progression, the consequence of HIV-1 infection for HLA-I/KIR interactions remain largely unknown.
Design: We determined changes in HLA-I presented peptides resulting from HIV-1-infection of primary human CD4 T cells and assessed the impact of changes in peptide repertoires on HLA-I/KIR interactions.
Methods: Liquid chromatography-coupled tandem mass spectrometry to identify HLA-I presented peptides, cell-based in-vitro assays to evaluate functional consequences of alterations in immunopeptidome and atomistic molecular dynamics simulations to confirm experimental data.
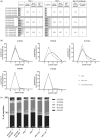
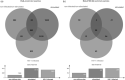
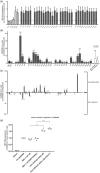
Results: A total of 583 peptides exclusively presented on HIV-1-infected cells were identified, of which only 0.2% represented HIV-1 derived peptides. Focusing on HLA-C*03 : 04/KIR2DL3 interactions, we observed that HLA-C*03 : 04-presented peptides derived from noninfected CD4 T cells mediated stronger binding of inhibitory KIR2DL3 than peptides derived from HIV-1-infected cells. Furthermore, the most abundant peptide presented by HLA-C*03 : 04 on noninfected CD4 T cells (VIYPARISL) mediated the strongest KIR2DL3-binding, while the most abundant peptide presented on HIV-1-infected cells (YAIQATETL) did not mediate KIR2DL3-binding. Molecular dynamics simulations of HLA-C*03 : 04/KIR2DL3 interactions in the context of these two peptides revealed that VIYPARISL significantly enhanced the HLA-C*03 : 04/peptide contact area to KIR2DL3 compared with YAIQATETL.
Conclusion: These data demonstrate that HIV-1 infection-induced changes in HLA-I-presented peptides can reduce engagement of inhibitory KIRs, providing a mechanism for enhanced activation of NK cells by virus-infected cells.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures


Comment in
-
Regulation of natural killer cells: analog peptide handshake goes digital.AIDS. 2020 Oct 1;34(12):1857-1858. doi: 10.1097/QAD.0000000000002657. AIDS. 2020. PMID: 32889856 No abstract available.
Similar articles
-
Sequence variations in HCV core-derived epitopes alter binding of KIR2DL3 to HLA-C∗03:04 and modulate NK cell function.J Hepatol. 2016 Aug;65(2):252-8. doi: 10.1016/j.jhep.2016.03.016. Epub 2016 Apr 4. J Hepatol. 2016. PMID: 27057987 Free PMC article.
-
The Education of NK Cells Determines Their Responsiveness to Autologous HIV-Infected CD4 T Cells.J Virol. 2019 Nov 13;93(23):e01185-19. doi: 10.1128/JVI.01185-19. Print 2019 Dec 1. J Virol. 2019. PMID: 31511383 Free PMC article.
-
Selection of an HLA-C*03:04-Restricted HIV-1 p24 Gag Sequence Variant Is Associated with Viral Escape from KIR2DL3+ Natural Killer Cells: Data from an Observational Cohort in South Africa.PLoS Med. 2015 Nov 17;12(11):e1001900; discussion e1001900. doi: 10.1371/journal.pmed.1001900. eCollection 2015 Nov. PLoS Med. 2015. PMID: 26575988 Free PMC article.
-
The role genes encoding of killer cell immunoglobulin-like receptors (KIRs) and their ligands in susceptibility to and progression of HIV infection.Postepy Hig Med Dosw (Online). 2016 Dec 31;70(0):1409-1423. doi: 10.5604/17322693.1227771. Postepy Hig Med Dosw (Online). 2016. PMID: 28100849 Review.
-
Missing or altered self: human NK cell receptors that recognize HLA-C.Immunogenetics. 2017 Aug;69(8-9):567-579. doi: 10.1007/s00251-017-1001-y. Epub 2017 Jul 10. Immunogenetics. 2017. PMID: 28695291 Free PMC article. Review.
Cited by
-
The New Kid on the Block: HLA-C, a Key Regulator of Natural Killer Cells in Viral Immunity.Cells. 2021 Nov 10;10(11):3108. doi: 10.3390/cells10113108. Cells. 2021. PMID: 34831331 Free PMC article. Review.
-
Targeting NK Cells for HIV-1 Treatment and Reservoir Clearance.Front Immunol. 2022 Mar 16;13:842746. doi: 10.3389/fimmu.2022.842746. eCollection 2022. Front Immunol. 2022. PMID: 35371060 Free PMC article. Review.
-
Biogenesis of HLA Ligand Presentation in Immune Cells Upon Activation Reveals Changes in Peptide Length Preference.Front Immunol. 2020 Aug 28;11:1981. doi: 10.3389/fimmu.2020.01981. eCollection 2020. Front Immunol. 2020. PMID: 32983136 Free PMC article.
-
HLA-C Peptide Repertoires as Predictors of Clinical Response during Early SARS-CoV-2 Infection.Life (Basel). 2024 Sep 19;14(9):1181. doi: 10.3390/life14091181. Life (Basel). 2024. PMID: 39337964 Free PMC article.
-
Host KIR/HLA-C Genotypes Determine HIV-Mediated Changes of the NK Cell Repertoire and Are Associated With Vpu Sequence Variations Impacting Downmodulation of HLA-C.Front Immunol. 2022 Jul 15;13:922252. doi: 10.3389/fimmu.2022.922252. eCollection 2022. Front Immunol. 2022. PMID: 35911762 Free PMC article.
References
-
- Gleimer M, Parham P. Stress management: MHC class I and class I-like molecules as reporters of cellular stress. Immunity 2003; 19:469–477.. - PubMed
-
- Burrows SR, Rossjohn J, McCluskey J. Have we cut ourselves too short in mapping CTL epitopes?. Trends Immunol 2006; 27:11–16.. - PubMed
-
- Patil C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol 2001; 13:349–355.. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

