Filoviruses Use the HOPS Complex and UVRAG To Traffic to Niemann-Pick C1 Compartments during Viral Entry
- PMID: 32493822
- PMCID: PMC7394885
- DOI: 10.1128/JVI.01002-20
Filoviruses Use the HOPS Complex and UVRAG To Traffic to Niemann-Pick C1 Compartments during Viral Entry
Abstract
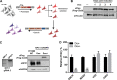
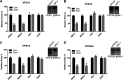
Ebola virus (EBOV) entry requires internalization into host cells and extensive trafficking through the endolysosomal network in order to reach late endosomal/lysosomal compartments that contain triggering factors for viral membrane fusion. These triggering factors include low-pH-activated cellular cathepsin proteases, which cleave the EBOV glycoprotein (GP), exposing a domain which binds to the filoviral receptor, the cholesterol transporter Niemann-Pick C1 (NPC1). Here, we report that trafficking of EBOV to NPC1 requires expression of the homotypic fusion and protein sorting (HOPS) tethering complex as well as its regulator, UV radiation resistance-associated gene (UVRAG). Using an inducible clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system, we demonstrated that depletion of HOPS subunits as well as UVRAG impairs entry by all pathogenic filoviruses. UVRAG depletion resulted in reduced delivery of EBOV virions to NPC1+ cellular compartments. Furthermore, we show that deletion of a domain on UVRAG known to be required for interaction with the HOPS complex results in impaired EBOV entry. Taken together, our studies demonstrate that EBOV requires both expression of and coordination between the HOPS complex and UVRAG in order to mediate efficient viral entry.IMPORTANCE Ebola viruses (EBOV) and other filoviruses cause sporadic and unpredictable outbreaks of highly lethal diseases. The lack of FDA-approved therapeutics, particularly ones with panfiloviral specificity, highlights the need for continued research efforts to understand aspects of the viral life cycle that are common to all filoviruses. As such, viral entry is of particular interest, as all filoviruses must reach cellular compartments containing the viral receptor Niemann-Pick C1 to enter cells. Here, we present an inducible CRISPR/Cas9 method to rapidly and efficiently generate knockout cells in order to interrogate the roles of a broad range of host factors in viral entry. Using this approach, we showed that EBOV entry depends on both the homotypic fusion and protein sorting (HOPS) tethering complex in coordination with UV radiation resistance-associated gene (UVRAG). Importantly, we demonstrate that the HOPS complex and UVRAG are required by all pathogenic filoviruses, representing potential targets for panfiloviral therapeutics.
Keywords: Ebola virus; filovirus; vesicular trafficking; virus entry.
Copyright © 2020 Bo et al.
Figures

Similar articles
-
Ebola virus entry requires the cholesterol transporter Niemann-Pick C1.Nature. 2011 Aug 24;477(7364):340-3. doi: 10.1038/nature10348. Nature. 2011. PMID: 21866103 Free PMC article.
-
Direct Intracellular Visualization of Ebola Virus-Receptor Interaction by In Situ Proximity Ligation.mBio. 2021 Jan 12;12(1):e03100-20. doi: 10.1128/mBio.03100-20. mBio. 2021. PMID: 33436438 Free PMC article.
-
Direct Visualization of Ebola Virus Fusion Triggering in the Endocytic Pathway.mBio. 2016 Feb 9;7(1):e01857-15. doi: 10.1128/mBio.01857-15. mBio. 2016. PMID: 26861015 Free PMC article.
-
Potential pharmacological strategies targeting the Niemann-Pick C1 receptor and Ebola virus glycoprotein interaction.Eur J Med Chem. 2021 Nov 5;223:113654. doi: 10.1016/j.ejmech.2021.113654. Epub 2021 Jun 19. Eur J Med Chem. 2021. PMID: 34175537 Review.
-
Filovirus entry: a novelty in the viral fusion world.Viruses. 2012 Feb;4(2):258-75. doi: 10.3390/v4020258. Epub 2012 Feb 7. Viruses. 2012. PMID: 22470835 Free PMC article. Review.
Cited by
-
Genome-wide CRISPR/Cas9 screen identifies SLC39A9 and PIK3C3 as crucial entry factors for Ebola virus infection.PLoS Pathog. 2024 Aug 22;20(8):e1012444. doi: 10.1371/journal.ppat.1012444. eCollection 2024 Aug. PLoS Pathog. 2024. PMID: 39173055 Free PMC article.
-
Ebola virus triggers receptor tyrosine kinase-dependent signaling to promote the delivery of viral particles to entry-conducive intracellular compartments.PLoS Pathog. 2021 Jan 29;17(1):e1009275. doi: 10.1371/journal.ppat.1009275. eCollection 2021 Jan. PLoS Pathog. 2021. PMID: 33513206 Free PMC article.
-
Navigating the Complex Landscape of Ebola Infection Treatment: A Review of Emerging Pharmacological Approaches.Infect Dis Ther. 2024 Jan;13(1):21-55. doi: 10.1007/s40121-023-00913-y. Epub 2024 Jan 19. Infect Dis Ther. 2024. PMID: 38240994 Free PMC article. Review.
-
Ebola Virus Entry Inhibitors.Adv Exp Med Biol. 2022;1366:155-170. doi: 10.1007/978-981-16-8702-0_10. Adv Exp Med Biol. 2022. PMID: 35412140
-
Molecular insights into the Ebola virus life cycle.Nat Microbiol. 2024 Jun;9(6):1417-1426. doi: 10.1038/s41564-024-01703-z. Epub 2024 May 23. Nat Microbiol. 2024. PMID: 38783022 Review.
References
-
- Henao-Restrepo AM, Camacho A, Longini IM, Watson CH, Edmunds WJ, Egger M, Carroll MW, Dean NE, Diatta I, Doumbia M, Draguez B, Duraffour S, Enwere G, Grais R, Gunther S, Gsell P-S, Hossmann S, Watle SV, Kondé MK, Kéïta S, Kone S, Kuisma E, Levine MM, Mandal S, Mauget T, Norheim G, Riveros X, Soumah A, Trelle S, Vicari AS, Røttingen J-A, Kieny M-P. 2017. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ca Suffit!). Lancet 389:505–518. doi:10.1016/S0140-6736(16)32621-6. - DOI - PMC - PubMed
-
- Qiu X, Audet J, Lv M, He S, Wong G, Wei H, Luo L, Fernando L, Kroeker A, Fausther Bovendo H, Bello A, Li F, Ye P, Jacobs M, Ippolito G, Saphire EO, Bi S, Shen B, Gao GF, Zeitlin L, Feng J, Zhang B, Kobinger GP. 2016. Two-mAb cocktail protects macaques against the Makona variant of Ebola virus. Sci Transl Med 8:329ra33. doi:10.1126/scitranslmed.aad9875. - DOI - PMC - PubMed
-
- Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, Johnson A, Morton J, Swope K, Bohorov O, Bohorova N, Goodman C, Kim D, Pauly MH, Velasco J, Pettitt J, Olinger GG, Whaley K, Xu B, Strong JE, Zeitlin L, Kobinger GP. 2014. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514:47–53. doi:10.1038/nature13777. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

