Human norovirus targets enteroendocrine epithelial cells in the small intestine
- PMID: 32488028
- PMCID: PMC7265440
- DOI: 10.1038/s41467-020-16491-3
Human norovirus targets enteroendocrine epithelial cells in the small intestine
Abstract
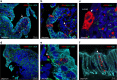
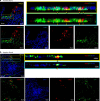
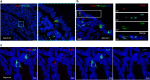
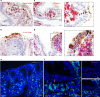
Human noroviruses are a major cause of diarrheal illness, but pathogenesis is poorly understood. Here, we investigate the cellular tropism of norovirus in specimens from four immunocompromised patients. Abundant norovirus antigen and RNA are detected throughout the small intestinal tract in jejunal and ileal tissue from one pediatric intestinal transplant recipient with severe gastroenteritis. Negative-sense viral RNA, a marker of active viral replication, is found predominantly in intestinal epithelial cells, with chromogranin A-positive enteroendocrine cells (EECs) identified as a permissive cell type in this patient. These findings are consistent with the detection of norovirus-positive EECs in the other three immunocompromised patients. Investigation of the signaling pathways induced in EECs that mediate communication between the gut and brain may clarify mechanisms of pathogenesis and lead to the development of in vitro model systems in which to evaluate norovirus vaccines and treatment.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Norovirus Replication in Human Intestinal Epithelial Cells Is Restricted by the Interferon-Induced JAK/STAT Signaling Pathway and RNA Polymerase II-Mediated Transcriptional Responses.mBio. 2020 Mar 17;11(2):e00215-20. doi: 10.1128/mBio.00215-20. mBio. 2020. PMID: 32184238 Free PMC article.
-
Lack of norovirus replication and histo-blood group antigen expression in 3-dimensional intestinal epithelial cells.Emerg Infect Dis. 2013 Mar;19(3):431-8. doi: 10.3201/eid1903.121029. Emerg Infect Dis. 2013. PMID: 23622517 Free PMC article.
-
Expression of Ifnlr1 on Intestinal Epithelial Cells Is Critical to the Antiviral Effects of Interferon Lambda against Norovirus and Reovirus.J Virol. 2017 Mar 13;91(7):e02079-16. doi: 10.1128/JVI.02079-16. Print 2017 Apr 1. J Virol. 2017. PMID: 28077655 Free PMC article.
-
Recent advances in understanding norovirus pathogenesis.J Med Virol. 2016 Nov;88(11):1837-43. doi: 10.1002/jmv.24559. Epub 2016 May 5. J Med Virol. 2016. PMID: 27110852 Free PMC article. Review.
-
Human norovirus transmission and evolution in a changing world.Nat Rev Microbiol. 2016 Jul;14(7):421-33. doi: 10.1038/nrmicro.2016.48. Epub 2016 May 23. Nat Rev Microbiol. 2016. PMID: 27211790 Review.
Cited by
-
A rhesus macaque model of human norovirus infection.Nat Microbiol. 2024 Mar;9(3):593-594. doi: 10.1038/s41564-023-01597-3. Nat Microbiol. 2024. PMID: 38378852 No abstract available.
-
Understanding neurotropic enteric viruses: routes of infection and mechanisms of attenuation.Cell Mol Life Sci. 2024 Oct 4;81(1):413. doi: 10.1007/s00018-024-05450-6. Cell Mol Life Sci. 2024. PMID: 39365457 Free PMC article. Review.
-
Human Norovirus Efficiently Replicates in Differentiated 3D-Human Intestinal Enteroids.J Virol. 2022 Nov 23;96(22):e0085522. doi: 10.1128/jvi.00855-22. Epub 2022 Nov 7. J Virol. 2022. PMID: 36342297 Free PMC article.
-
Experimental Methods to Study the Pathogenesis of Human Enteric RNA Viruses.Viruses. 2021 May 25;13(6):975. doi: 10.3390/v13060975. Viruses. 2021. PMID: 34070283 Free PMC article. Review.
-
Norovirus Intestinal Infection and Lewy Body Disease in an Older Patient with Acute Cognitive Impairment.Int J Mol Sci. 2022 Jul 29;23(15):8376. doi: 10.3390/ijms23158376. Int J Mol Sci. 2022. PMID: 35955510 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

