Beyond the coding genome: non-coding mutations and cancer
- PMID: 32472759
- PMCID: PMC7611228
- DOI: 10.2741/4879
Beyond the coding genome: non-coding mutations and cancer
Abstract
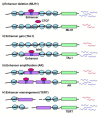
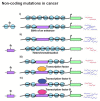
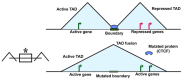
Latest advancements in genomics involving individuals from different races and geographical locations has led to the identification of thousands of common as well as rare genetic variants and copy number variations (CNVs). These studies have surprisingly revealed that the majority of genetic variation is not present within the coding region but rather in the non-coding region of the genome, which is also termed as "Medical Genome". This short review describes how mutations/variations within; regulatory sequences, architectural proteins and transcriptional regulators give rise to the aberrant gene expression profiles that drives cellular transformations and malignancies.
Figures
Similar articles
-
Cancer regulatory variation.Curr Opin Genet Dev. 2021 Feb;66:41-49. doi: 10.1016/j.gde.2020.11.010. Epub 2021 Jan 7. Curr Opin Genet Dev. 2021. PMID: 33422949 Review.
-
Tae Hoon Kim: knows his boundaries. Interview by Ruth Williams.J Cell Biol. 2008 Oct 6;183(1):4-5. doi: 10.1083/jcb.1831pi. J Cell Biol. 2008. PMID: 18838549 Free PMC article. No abstract available.
-
Identification and characterization of cis-regulatory elements 'insulator and repressor' in PPARD gene.Epigenomics. 2018 May;10(5):613-627. doi: 10.2217/epi-2017-0139. Epub 2018 Mar 27. Epigenomics. 2018. PMID: 29583017
-
Genomic approaches for the discovery of CFTR regulatory elements.Transcription. 2011 Jan-Feb;2(1):23-7. doi: 10.4161/trns.2.1.13693. Transcription. 2011. PMID: 21326906 Free PMC article.
-
Chromatin insulators and long-distance interactions in Drosophila.FEBS Lett. 2014 Jan 3;588(1):8-14. doi: 10.1016/j.febslet.2013.10.039. Epub 2013 Nov 5. FEBS Lett. 2014. PMID: 24211836 Review.
Cited by
-
Bioinformatic Evaluation of Features on Cis-regulatory Elements at 6q25.1.Bioinform Biol Insights. 2023 Apr 25;17:11779322231167971. doi: 10.1177/11779322231167971. eCollection 2023. Bioinform Biol Insights. 2023. PMID: 37124129 Free PMC article. Review.
-
Can Genetic Markers Predict the Sporadic Form of Alzheimer's Disease? An Updated Review on Genetic Peripheral Markers.Int J Mol Sci. 2023 Aug 30;24(17):13480. doi: 10.3390/ijms241713480. Int J Mol Sci. 2023. PMID: 37686283 Free PMC article. Review.
-
Effect of HPSE and HPSE2 SNPs on the Risk of Developing Primary Paraskeletal Multiple Myeloma.Cells. 2023 Mar 16;12(6):913. doi: 10.3390/cells12060913. Cells. 2023. PMID: 36980254 Free PMC article.
-
Pathophysiology of human hearing loss associated with variants in myosins.Front Physiol. 2024 Mar 18;15:1374901. doi: 10.3389/fphys.2024.1374901. eCollection 2024. Front Physiol. 2024. PMID: 38562617 Free PMC article. Review.
-
Androgen Receptor-Mediated Transcription in Prostate Cancer.Cells. 2022 Mar 5;11(5):898. doi: 10.3390/cells11050898. Cells. 2022. PMID: 35269520 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

