Structural Insights into APOBEC3-Mediated Lentiviral Restriction
- PMID: 32471198
- PMCID: PMC7354603
- DOI: 10.3390/v12060587
Structural Insights into APOBEC3-Mediated Lentiviral Restriction
Abstract
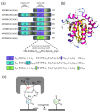
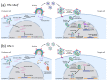
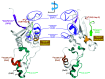
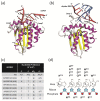
Mammals have developed clever adaptive and innate immune defense mechanisms to protect against invading bacterial and viral pathogens. Human innate immunity is continuously evolving to expand the repertoire of restriction factors and one such family of intrinsic restriction factors is the APOBEC3 (A3) family of cytidine deaminases. The coordinated expression of seven members of the A3 family of cytidine deaminases provides intrinsic immunity against numerous foreign infectious agents and protects the host from exogenous retroviruses and endogenous retroelements. Four members of the A3 proteins-A3G, A3F, A3H, and A3D-restrict HIV-1 in the absence of virion infectivity factor (Vif); their incorporation into progeny virions is a prerequisite for cytidine deaminase-dependent and -independent activities that inhibit viral replication in the host target cell. HIV-1 encodes Vif, an accessory protein that antagonizes A3 proteins by targeting them for polyubiquitination and subsequent proteasomal degradation in the virus producing cells. In this review, we summarize our current understanding of the role of human A3 proteins as barriers against HIV-1 infection, how Vif overcomes their antiviral activity, and highlight recent structural and functional insights into A3-mediated restriction of lentiviruses.
Keywords: APOBEC3G; CBFβ; Vif; cullin 5; cytidine deamination; elongin b and c; hypermutation; nucleic acid binding; proteasomal degradation; restriction factor; substrate selection.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
HIV-1 and HIV-2 Vif interact with human APOBEC3 proteins using completely different determinants.J Virol. 2014 Sep 1;88(17):9893-908. doi: 10.1128/JVI.01318-14. Epub 2014 Jun 18. J Virol. 2014. PMID: 24942576 Free PMC article.
-
Feline Immunodeficiency Virus Evolutionarily Acquires Two Proteins, Vif and Protease, Capable of Antagonizing Feline APOBEC3.J Virol. 2017 May 12;91(11):e00250-17. doi: 10.1128/JVI.00250-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28331087 Free PMC article.
-
Structural Features of Antiviral APOBEC3 Proteins are Linked to Their Functional Activities.Front Microbiol. 2011 Dec 21;2:258. doi: 10.3389/fmicb.2011.00258. eCollection 2011. Front Microbiol. 2011. PMID: 22203821 Free PMC article.
-
[APOBEC3s: history of an antiviral and mutagenic protein family].Virologie (Montrouge). 2020 Dec 1;24(6):381-418. doi: 10.1684/vir.2020.0870. Virologie (Montrouge). 2020. PMID: 33441290 Review. French.
-
Structural perspectives on HIV-1 Vif and APOBEC3 restriction factor interactions.Protein Sci. 2020 Feb;29(2):391-406. doi: 10.1002/pro.3729. Epub 2019 Nov 29. Protein Sci. 2020. PMID: 31518043 Free PMC article. Review.
Cited by
-
The structural basis for HIV-1 Vif antagonism of human APOBEC3G.Nature. 2023 Mar;615(7953):728-733. doi: 10.1038/s41586-023-05779-1. Epub 2023 Feb 8. Nature. 2023. PMID: 36754086 Free PMC article.
-
HPV16 and HPV18 Genome Structure, Expression, and Post-Transcriptional Regulation.Int J Mol Sci. 2022 Apr 29;23(9):4943. doi: 10.3390/ijms23094943. Int J Mol Sci. 2022. PMID: 35563334 Free PMC article. Review.
-
Deaminase-Independent Mode of Antiretroviral Action in Human and Mouse APOBEC3 Proteins.Microorganisms. 2020 Dec 12;8(12):1976. doi: 10.3390/microorganisms8121976. Microorganisms. 2020. PMID: 33322756 Free PMC article. Review.
-
Potential APOBEC-mediated RNA editing of the genomes of SARS-CoV-2 and other coronaviruses and its impact on their longer term evolution.Virology. 2021 Apr;556:62-72. doi: 10.1016/j.virol.2020.12.018. Epub 2021 Jan 7. Virology. 2021. PMID: 33545556 Free PMC article. Review.
-
Let It Go: HIV-1 cis-Acting Repressive Sequences.J Virol. 2021 Jul 12;95(15):e0034221. doi: 10.1128/JVI.00342-21. Epub 2021 Jul 12. J Virol. 2021. PMID: 33980600 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

