The domesticated transposase ALP2 mediates formation of a novel Polycomb protein complex by direct interaction with MSI1, a core subunit of Polycomb Repressive Complex 2 (PRC2)
- PMID: 32463832
- PMCID: PMC7282668
- DOI: 10.1371/journal.pgen.1008681
The domesticated transposase ALP2 mediates formation of a novel Polycomb protein complex by direct interaction with MSI1, a core subunit of Polycomb Repressive Complex 2 (PRC2)
Abstract
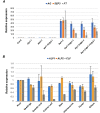
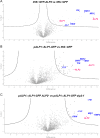
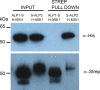
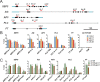
A large fraction of plant genomes is composed of transposable elements (TE), which provide a potential source of novel genes through "domestication"-the process whereby the proteins encoded by TE diverge in sequence, lose their ability to catalyse transposition and instead acquire novel functions for their hosts. In Arabidopsis, ANTAGONIST OF LIKE HETEROCHROMATIN PROTEIN 1 (ALP1) arose by domestication of the nuclease component of Harbinger class TE and acquired a new function as a component of POLYCOMB REPRESSIVE COMPLEX 2 (PRC2), a histone H3K27me3 methyltransferase involved in regulation of host genes and in some cases TE. It was not clear how ALP1 associated with PRC2, nor what the functional consequence was. Here, we identify ALP2 genetically as a suppressor of Polycomb-group (PcG) mutant phenotypes and show that it arose from the second, DNA binding component of Harbinger transposases. Molecular analysis of PcG compromised backgrounds reveals that ALP genes oppose silencing and H3K27me3 deposition at key PcG target genes. Proteomic analysis reveals that ALP1 and ALP2 are components of a variant PRC2 complex that contains the four core components but lacks plant-specific accessory components such as the H3K27me3 reader LIKE HETEROCHROMATION PROTEIN 1 (LHP1). We show that the N-terminus of ALP2 interacts directly with ALP1, whereas the C-terminus of ALP2 interacts with MULTICOPY SUPPRESSOR OF IRA1 (MSI1), a core component of PRC2. Proteomic analysis reveals that in alp2 mutant backgrounds ALP1 protein no longer associates with PRC2, consistent with a role for ALP2 in recruitment of ALP1. We suggest that the propensity of Harbinger TE to insert in gene-rich regions of the genome, together with the modular two component nature of their transposases, has predisposed them for domestication and incorporation into chromatin modifying complexes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Kicking against the PRCs - A Domesticated Transposase Antagonises Silencing Mediated by Polycomb Group Proteins and Is an Accessory Component of Polycomb Repressive Complex 2.PLoS Genet. 2015 Dec 7;11(12):e1005660. doi: 10.1371/journal.pgen.1005660. eCollection 2015 Dec. PLoS Genet. 2015. PMID: 26642436 Free PMC article.
-
Arabidopsis MSI1 connects LHP1 to PRC2 complexes.EMBO J. 2013 Jul 17;32(14):2073-85. doi: 10.1038/emboj.2013.145. Epub 2013 Jun 18. EMBO J. 2013. PMID: 23778966 Free PMC article.
-
H2A monoubiquitination in Arabidopsis thaliana is generally independent of LHP1 and PRC2 activity.Genome Biol. 2017 Apr 12;18(1):69. doi: 10.1186/s13059-017-1197-z. Genome Biol. 2017. PMID: 28403905 Free PMC article.
-
Tissue-Specific Tumour Suppressor and Oncogenic Activities of the Polycomb-like Protein MTF2.Genes (Basel). 2023 Sep 27;14(10):1879. doi: 10.3390/genes14101879. Genes (Basel). 2023. PMID: 37895228 Free PMC article. Review.
-
Polycomb group and trithorax group proteins in Arabidopsis.Biochim Biophys Acta. 2007 May-Jun;1769(5-6):375-82. doi: 10.1016/j.bbaexp.2007.01.010. Epub 2007 Feb 7. Biochim Biophys Acta. 2007. PMID: 17363079 Review.
Cited by
-
Recurrent co-domestication of PIF/Harbinger transposable element proteins in insects.Mob DNA. 2022 Nov 30;13(1):28. doi: 10.1186/s13100-022-00282-2. Mob DNA. 2022. PMID: 36451208 Free PMC article.
-
Polycomb Repressive Complex 2 in Eukaryotes-An Evolutionary Perspective.Epigenomes. 2022 Jan 17;6(1):3. doi: 10.3390/epigenomes6010003. Epigenomes. 2022. PMID: 35076495 Free PMC article. Review.
-
Diversity of Harbinger-like Transposons in Teleost Fish Genomes.Animals (Basel). 2022 May 31;12(11):1429. doi: 10.3390/ani12111429. Animals (Basel). 2022. PMID: 35681893 Free PMC article.
-
Transposase expression, element abundance, element size, and DNA repair determine the mobility and heritability of PIF/Pong/Harbinger transposable elements.Front Cell Dev Biol. 2023 Jun 9;11:1184046. doi: 10.3389/fcell.2023.1184046. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37363729 Free PMC article.
-
Dynamics of H3K27me3 Modification on Plant Adaptation to Environmental Cues.Plants (Basel). 2021 Jun 8;10(6):1165. doi: 10.3390/plants10061165. Plants (Basel). 2021. PMID: 34201297 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

