Lung Tumor Cell-Derived Exosomes Promote M2 Macrophage Polarization
- PMID: 32456301
- PMCID: PMC7290460
- DOI: 10.3390/cells9051303
Lung Tumor Cell-Derived Exosomes Promote M2 Macrophage Polarization
Abstract
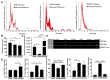
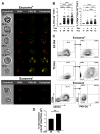
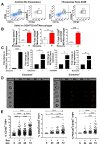
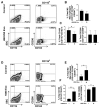
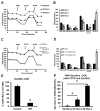
Cellular cross-talk within the tumor microenvironment (TME) by exosomes is known to promote tumor progression. Tumor promoting macrophages with an M2 phenotype are suppressors of anti-tumor immunity. However, the impact of tumor-derived exosomes in modulating macrophage polarization in the lung TME is largely unknown. Herein, we investigated if lung tumor-derived exosomes alter transcriptional and bioenergetic signatures of M0 macrophages and polarize them to an M2 phenotype. The concentration of exosomes produced by p53 null H358 lung tumor cells was significantly reduced compared to A549 (p53 wild-type) lung tumor cells, consistent with p53-mediated regulation of exosome production. In co-culture studies, M0 macrophages internalized tumor-derived exosomes, and differentiated into M2 phenotype. Importantly, we demonstrate that tumor-derived exosomes enhance the oxygen consumption rate of macrophages, altering their bioenergetic state consistent with that of M2 macrophages. In vitro co-cultures of M0 macrophages with H358 exosomes demonstrated that exosome-induced M2 polarization may be p53 independent. Murine bone marrow cells and bone marrow-derived myeloid-derived suppressor cells (MDSCs) co-cultured with lewis lung carcinoma (LLC)-derived exosomes differentiated to M2 macrophages. Collectively, these studies provide evidence for a novel role for lung tumor-exosomes in M2 macrophage polarization, which then offers new therapeutic targets for immunotherapy of lung cancer.
Keywords: M2 macrophages; exosomes; lung cancer; macrophage polarization; myeloid-derived suppressor cells; tumor associated macrophages.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

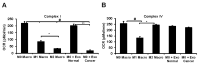
Similar articles
-
Hypoxic lung cancer cell-derived exosomal miR-21 mediates macrophage M2 polarization and promotes cancer cell proliferation through targeting IRF1.World J Surg Oncol. 2022 Jul 27;20(1):241. doi: 10.1186/s12957-022-02706-y. World J Surg Oncol. 2022. PMID: 35897096 Free PMC article.
-
Diffuse large B-cell lymphoma-derived exosomes push macrophage polarization toward M2 phenotype via GP130/STAT3 signaling pathway.Chem Biol Interact. 2022 Jan 25;352:109779. doi: 10.1016/j.cbi.2021.109779. Epub 2021 Dec 17. Chem Biol Interact. 2022. PMID: 34922904
-
Oral squamous cell carcinoma-derived exosomes promote M2 subtype macrophage polarization mediated by exosome-enclosed miR-29a-3p.Am J Physiol Cell Physiol. 2019 May 1;316(5):C731-C740. doi: 10.1152/ajpcell.00366.2018. Epub 2019 Feb 27. Am J Physiol Cell Physiol. 2019. PMID: 30811223
-
Tumor-derived exosomes in the regulation of macrophage polarization.Inflamm Res. 2020 May;69(5):435-451. doi: 10.1007/s00011-020-01318-0. Epub 2020 Mar 11. Inflamm Res. 2020. PMID: 32162012 Review.
-
[Influencing Factors and Significance of Tumor-associated Macrophage Polarization in Tumor Microenvironment].Zhongguo Fei Ai Za Zhi. 2023 Mar 20;26(3):228-237. doi: 10.3779/j.issn.1009-3419.2023.106.07. Zhongguo Fei Ai Za Zhi. 2023. PMID: 37035885 Free PMC article. Review. Chinese.
Cited by
-
Establish a novel immune-related gene prognostic risk index (IRGPRI) associated with CD8+ cytotoxic T lymphocytes in non-small-cell lung cancer (NSCLC).Heliyon. 2024 Sep 24;10(19):e38324. doi: 10.1016/j.heliyon.2024.e38324. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39397989 Free PMC article.
-
Tumour-associated macrophages heterogeneity drives resistance to clinical therapy.Expert Rev Mol Med. 2022 Apr 11;24:e17. doi: 10.1017/erm.2022.8. Expert Rev Mol Med. 2022. PMID: 35400355 Free PMC article. Review.
-
Tumor cell-derived exosomal microRNA-146a promotes non-small cell lung cancer cell invasion and proliferation by inhibiting M1 macrophage polarization.Ann Transl Med. 2022 Dec;10(24):1307. doi: 10.21037/atm-22-5565. Ann Transl Med. 2022. PMID: 36660623 Free PMC article.
-
Tumor-derived exosomes: immune properties and clinical application in lung cancer.Cancer Drug Resist. 2022 Feb 8;5(1):102-113. doi: 10.20517/cdr.2021.99. eCollection 2022. Cancer Drug Resist. 2022. PMID: 35582534 Free PMC article. Review.
-
Systematic Construction and Validation of a Novel Macrophage Differentiation-Associated Prognostic Model for Clear Cell Renal Cell Carcinoma.Front Genet. 2022 Jun 14;13:877656. doi: 10.3389/fgene.2022.877656. eCollection 2022. Front Genet. 2022. PMID: 35774505 Free PMC article.
References
-
- Cancer Facts and Figures. [(accessed on 1 April 2020)];2017 Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts....
-
- World Health Organization . The Global Burden of Disease: 2004 Update. World Health Organization; Geneva, Switzerland: 2004.
-
- Woo E.Y., Chu C.S., Goletz T.J., Schlienger K., Yeh H., Coukos G., Rubin S.C., Kaiser L.R., June C.H. Regulatory CD4(+)CD25(+) T cells in tumors from patients with early-stage non-small cell lung cancer and late-stage ovarian cancer. Cancer Res. 2001;61:4766–4772. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

