A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping
- PMID: 32443810
- PMCID: PMC7291026
- DOI: 10.3390/cells9051267
A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping
Abstract
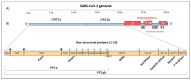
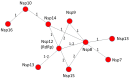
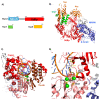
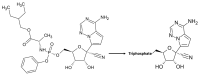
The current coronavirus disease-2019 (COVID-19) pandemic is due to the novel coronavirus SARS-CoV-2. The scientific community has mounted a strong response by accelerating research and innovation, and has quickly set the foundation for understanding the molecular determinants of the disease for the development of targeted therapeutic interventions. The replication of the viral genome within the infected cells is a key stage of the SARS-CoV-2 life cycle. It is a complex process involving the action of several viral and host proteins in order to perform RNA polymerization, proofreading and final capping. This review provides an update of the structural and functional data on the key actors of the replicatory machinery of SARS-CoV-2, to fill the gaps in the currently available structural data, which is mainly obtained through homology modeling. Moreover, learning from similar viruses, we collect data from the literature to reconstruct the pattern of interactions among the protein actors of the SARS-CoV-2 RNA polymerase machinery. Here, an important role is played by co-factors such as Nsp8 and Nsp10, not only as allosteric activators but also as molecular connectors that hold the entire machinery together to enhance the efficiency of RNA replication.
Keywords: COVID19; RNA replication; SARS-CoV-2; infectious disease; protein structure.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin.Nat Commun. 2020 Jul 24;11(1):3717. doi: 10.1038/s41467-020-17495-9. Nat Commun. 2020. PMID: 32709887 Free PMC article.
-
The Enzymatic Activity of the nsp14 Exoribonuclease Is Critical for Replication of MERS-CoV and SARS-CoV-2.J Virol. 2020 Nov 9;94(23):e01246-20. doi: 10.1128/JVI.01246-20. Print 2020 Nov 9. J Virol. 2020. PMID: 32938769 Free PMC article.
-
Structural basis of RNA cap modification by SARS-CoV-2.Nat Commun. 2020 Jul 24;11(1):3718. doi: 10.1038/s41467-020-17496-8. Nat Commun. 2020. PMID: 32709886 Free PMC article.
-
The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19.Protein J. 2020 Jun;39(3):198-216. doi: 10.1007/s10930-020-09901-4. Protein J. 2020. PMID: 32447571 Free PMC article. Review.
-
Replication of the coronavirus genome: A paradox among positive-strand RNA viruses.J Biol Chem. 2022 May;298(5):101923. doi: 10.1016/j.jbc.2022.101923. Epub 2022 Apr 10. J Biol Chem. 2022. PMID: 35413290 Free PMC article. Review.
Cited by
-
Is a healthy microbiome responsible for lower mortality in COVID-19?Biologia (Bratisl). 2021;76(2):819-829. doi: 10.2478/s11756-020-00614-8. Epub 2020 Oct 15. Biologia (Bratisl). 2021. PMID: 33078028 Free PMC article.
-
Insight into genomic organization of pathogenic coronaviruses, SARS-CoV-2: Implication for emergence of new variants, laboratory diagnosis and treatment options.Front Mol Med. 2022 Oct 17;2:917201. doi: 10.3389/fmmed.2022.917201. eCollection 2022. Front Mol Med. 2022. PMID: 39157715 Free PMC article. Review.
-
Molecular Epidemiology Analysis of SARS-CoV-2 Strains Circulating in Romania during the First Months of the Pandemic.Life (Basel). 2020 Aug 14;10(8):152. doi: 10.3390/life10080152. Life (Basel). 2020. PMID: 32823907 Free PMC article.
-
Antisense technology as a potential strategy for the treatment of coronaviruses infection: With focus on COVID-19.IET Nanobiotechnol. 2022 May;16(3):67-77. doi: 10.1049/nbt2.12079. Epub 2022 Mar 10. IET Nanobiotechnol. 2022. PMID: 35274474 Free PMC article. Review.
-
The Complexity of SARS-CoV-2 Infection and the COVID-19 Pandemic.Front Microbiol. 2022 Feb 10;13:789882. doi: 10.3389/fmicb.2022.789882. eCollection 2022. Front Microbiol. 2022. PMID: 35222327 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

