Potential role of miR-155-5p in fat deposition and skeletal muscle development of chicken
- PMID: 32441300
- PMCID: PMC7269915
- DOI: 10.1042/BSR20193796
Potential role of miR-155-5p in fat deposition and skeletal muscle development of chicken
Abstract
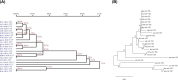
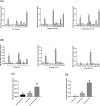
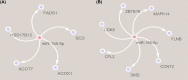
miR-155 has multiple functions in many physiological and pathological processes. However, little is known about the expression characteristics of avian miR-155. In the present study, partial pri-miR-155 sequences were cloned from AA+ broiler, Sanhuang broiler and Hy-Line Brown layer, respectively. Stem-loop qRT-PCR was performed to detect the miR-155-5p spatiotemporal expression profiles of each chicken breed, and the target genes of miR-155-5p were predicted in Gene Oncology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The results showed that the partial pri-miR-155 sequences of different breeds of chicken were high conserved. The expression patterns of miR-155-5p between broiler and layer were basically similar, and miR-155-5p was expressed highly in immune related tissues (spleen, thymus and bursa). In the same old chicken (14 days old), miR-155-5p expression activity of fat tissue all had higher level in the three chicken breeds, but the expression activities in skeletal muscle of broilers were significantly lower than that of layer (P<0.05). In different development stages of Hy-Line Brown layer, miR-155-5p expression activities in skeletal muscle of 14-day-old and 10-month-old layers were significantly lower than that of 24-month-old layer (P<0.05). Fat related target genes (ACOX1, ACOT7, FADS1, SCD and HSD17B12) and skeletal muscle related target genes (CCNT2, DMD, CFL2, MAPK14, FLNB, ZBTB18 and CDK5) of miR-155-5p were predicted, respectively. The results indicate that miR-155-5p may be an important factor inhibiting the fat deposition and skeletal muscle development in chicken.
Keywords: chicken; clone; expression; fat; miR-155-5p; skeletal muscle.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures
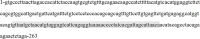
Similar articles
-
MicroRNA profiling associated with muscle growth in modern broilers compared to an unselected chicken breed.BMC Genomics. 2018 Sep 17;19(1):683. doi: 10.1186/s12864-018-5061-7. BMC Genomics. 2018. PMID: 30223794 Free PMC article.
-
Differential expression profile of microRNA in yak skeletal muscle and adipose tissue during development.Genes Genomics. 2020 Nov;42(11):1347-1359. doi: 10.1007/s13258-020-00988-8. Epub 2020 Sep 29. Genes Genomics. 2020. PMID: 32996042
-
Systematic transcriptome-wide analysis of mRNA-miRNA interactions reveals the involvement of miR-142-5p and its target (FOXO3) in skeletal muscle growth in chickens.Mol Genet Genomics. 2018 Feb;293(1):69-80. doi: 10.1007/s00438-017-1364-7. Epub 2017 Sep 2. Mol Genet Genomics. 2018. PMID: 28866851
-
A systematic analysis of the skeletal muscle miRNA transcriptome of chicken varieties with divergent skeletal muscle growth identifies novel miRNAs and differentially expressed miRNAs.BMC Genomics. 2011 Apr 13;12:186. doi: 10.1186/1471-2164-12-186. BMC Genomics. 2011. PMID: 21486491 Free PMC article.
-
Transcriptome landscapes of differentially expressed genes related to fat deposits in Nandan-Yao chicken.Funct Integr Genomics. 2021 Jan;21(1):113-124. doi: 10.1007/s10142-020-00764-7. Epub 2021 Jan 6. Funct Integr Genomics. 2021. PMID: 33404913
Cited by
-
CircRNAs Related to Breast Muscle Development and Their Interaction Regulatory Network in Gushi Chicken.Genes (Basel). 2022 Oct 29;13(11):1974. doi: 10.3390/genes13111974. Genes (Basel). 2022. PMID: 36360215 Free PMC article.
-
Genome-wide detection of genetic structure and runs of homozygosity analysis in Anhui indigenous and Western commercial pig breeds using PorcineSNP80k data.BMC Genomics. 2022 May 17;23(1):373. doi: 10.1186/s12864-022-08583-9. BMC Genomics. 2022. PMID: 35581549 Free PMC article.
-
Genome-Wide Detection of Copy Number Variations and Evaluation of Candidate Copy Number Polymorphism Genes Associated With Complex Traits of Pigs.Front Vet Sci. 2022 Jun 30;9:909039. doi: 10.3389/fvets.2022.909039. eCollection 2022. Front Vet Sci. 2022. PMID: 35847642 Free PMC article.
-
Comparative Analysis of miRNA Expression Profiles in Skeletal Muscle of Bian Chickens at Different Embryonic Ages.Animals (Basel). 2022 Apr 13;12(8):1003. doi: 10.3390/ani12081003. Animals (Basel). 2022. PMID: 35454249 Free PMC article.
-
Comparative analysis of the growth differences between hybrid Ningdu Yellow chickens and their parentals.Poult Sci. 2024 Dec;103(12):104239. doi: 10.1016/j.psj.2024.104239. Epub 2024 Aug 22. Poult Sci. 2024. PMID: 39454533 Free PMC article.
References
-
- Bartel D.P. (2007) MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 131, 11–29 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

