Discovery of a potent SCAP degrader that ameliorates HFD-induced obesity, hyperlipidemia and insulin resistance via an autophagy-independent lysosomal pathway
- PMID: 32432943
- PMCID: PMC8354609
- DOI: 10.1080/15548627.2020.1757955
Discovery of a potent SCAP degrader that ameliorates HFD-induced obesity, hyperlipidemia and insulin resistance via an autophagy-independent lysosomal pathway
Abstract
SCAP (SREBF chaperone) regulates SREBFs (sterol regulatory element binding transcription factors) processing and stability, and, thus, becomes an emerging drug target to treat dyslipidemia and fatty liver disease. However, the current known SCAP inhibitors, such as oxysterols, induce endoplasmic reticulum (ER) stress and NR1H3/LXRα (nuclear receptor subfamily 1 group H member 3)-SREBF1/SREBP-1 c-mediated hepatic steatosis, which severely limited the clinical application of this inhibitor. In this study, we identified a small molecule, lycorine, which binds to SCAP, which suppressed the SREBF pathway without inducing ER stress or activating NR1H3. Mechanistically, lycorine promotes SCAP lysosomal degradation in a macroautophagy/autophagy-independent pathway, a mechanism completely distinct from current SCAP inhibitors. Furthermore, we determined that SQSTM1 captured SCAP after its exit from the ER. The interaction of SCAP and SQSTM1 requires the WD40 domain of SCAP and the TB domain of SQSTM1. Interestingly, lycorine triggers the lysosome translocation of SCAP independent of autophagy. We termed this novel protein degradation pathway as the SQSTM1-mediated autophagy-independent lysosomal degradation (SMAILD) pathway. In vivo, lycorine ameliorates high-fat diet-induced hyperlipidemia, hepatic steatosis, and insulin resistance in mice. Our study demonstrated that the inhibition of SCAP through the SMAILD pathway could be employed as a useful therapeutic strategy for treating metabolic diseases.Abbreviation: 25-OHD: 25-hydroxyvitamin D; 3-MA: 3-methyladenine; ABCG5: ATP binding cassette subfamily G member 5; ABCG8: ATP binding cassette subfamily G member 8; ACACA: acetyl-CoA carboxylase alpha; AEBSF: 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride; AHI: anhydroicaritin; AKT/protein kinase B: AKT serine/threonine kinase; APOE: apolipoprotein E; ATF6: activating transcription factor 6; ATG: autophagy-related; BAT: brown adipose tissue; CD274/PD-L1: CD274 molecule; CETSA: cellular thermal shift assay; CMA: chaperone-mediated autophagy; COPII: cytoplasmic coat protein complex-II; CQ: chloroquine; DDIT3/CHOP: DNA damage inducible transcript 3; DNL: de novo lipogenesis; EE: energy expenditure; EGFR: epithelial growth factor receptor; eMI: endosomal microautophagy; ERN1/IRE1α: endoplasmic reticulum to nucleus signaling 1; FADS2: fatty acid desaturase 2; FASN: fatty acid synthase; GOT1/AST: glutamic-oxaloacetic transaminase 1; GPT/ALT: glutamic-pyruvate transaminase; HMGCR: 3-hydroxy-3-methylglutaryl-CoA reductase; HMGCS1: 3-hydroxy-3-methylglutaryl-CoA synthase 1; HSP90B1/GRP94: heat shock protein 90 beta family member 1; HSPA5/GRP78: heat hock protein family A (Hsp70) member 5; HSPA8/HSC70: heat shock protein family A (Hsp70) member 8; INSIG1: insulin induced gene 1; LAMP2A: lysosomal associated membrane protein 2A; LDLR: low density lipoprotein receptor; LyTACs: lysosome targeting chimeras; MAP1LC3B/LC3B: microtubule associated protein 1 light chain 3 beta; MBTPS1: membrane bound transcription factor peptidase, site 1; MEF: mouse embryonic fibroblast; MST: microscale thermophoresis; MTOR: mechanistic target of rapamycin kinase; MVK: mevalonate kinase; PROTAC: proteolysis targeting chimera; RQ: respiratory quotient; SCAP: SREBF chaperone; SCD1: stearoyl-coenzemy A desaturase 1; SMAILD: sequestosome 1 mediated autophagy-independent lysosomal degradation; SQSTM1: sequestosome 1; SREBF: sterol regulatory element binding transcription factor; TNFRSF10B/DR5: TNF receptor superfamily member 10b; TRAF6: TNF receptor associated factor 6; UPR: unfolded protein response; WAT: white adipose tissue; XBP1: X-box binding protein 1.
Keywords: Autophagy; ER stress; SCAP; SQSTM1; SREBFs; lycorine.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
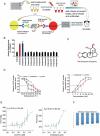
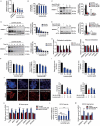

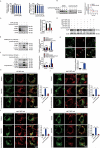
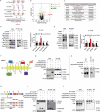
Similar articles
-
The ménage à trois of autophagy, lipid droplets and liver disease.Autophagy. 2022 Jan;18(1):50-72. doi: 10.1080/15548627.2021.1895658. Epub 2021 Apr 2. Autophagy. 2022. PMID: 33794741 Free PMC article. Review.
-
Autophagy in the physiological endometrium and cancer.Autophagy. 2021 May;17(5):1077-1095. doi: 10.1080/15548627.2020.1752548. Epub 2020 May 13. Autophagy. 2021. PMID: 32401642 Free PMC article. Review.
-
The GST-BHMT assay reveals a distinct mechanism underlying proteasome inhibition-induced macroautophagy in mammalian cells.Autophagy. 2015;11(5):812-32. doi: 10.1080/15548627.2015.1034402. Autophagy. 2015. PMID: 25984893 Free PMC article.
-
How autophagy controls the intestinal epithelial barrier.Autophagy. 2022 Jan;18(1):86-103. doi: 10.1080/15548627.2021.1909406. Epub 2021 Apr 27. Autophagy. 2022. PMID: 33906557 Free PMC article. Review.
-
TXNIP/VDUP1 attenuates steatohepatitis via autophagy and fatty acid oxidation.Autophagy. 2021 Sep;17(9):2549-2564. doi: 10.1080/15548627.2020.1834711. Epub 2020 Nov 16. Autophagy. 2021. PMID: 33190588 Free PMC article.
Cited by
-
The Art of Finding the Right Drug Target: Emerging Methods and Strategies.Pharmacol Rev. 2024 Aug 15;76(5):896-914. doi: 10.1124/pharmrev.123.001028. Pharmacol Rev. 2024. PMID: 38866560 Free PMC article. Review.
-
A cholesterol-binding bacterial toxin provides a strategy for identifying a specific Scap inhibitor that blocks lipid synthesis in animal cells.Proc Natl Acad Sci U S A. 2024 Feb 13;121(7):e2318024121. doi: 10.1073/pnas.2318024121. Epub 2024 Feb 8. Proc Natl Acad Sci U S A. 2024. PMID: 38330014 Free PMC article.
-
SREBPs as the potential target for solving the polypharmacy dilemma.Front Physiol. 2024 Jan 10;14:1272540. doi: 10.3389/fphys.2023.1272540. eCollection 2023. Front Physiol. 2024. PMID: 38269061 Free PMC article. Review.
-
A novel marine-derived anti-acute kidney injury agent targeting peroxiredoxin 1 and its nanodelivery strategy based on ADME optimization.Acta Pharm Sin B. 2024 Jul;14(7):3232-3250. doi: 10.1016/j.apsb.2024.03.005. Epub 2024 Mar 8. Acta Pharm Sin B. 2024. PMID: 39027260 Free PMC article.
-
Lycorine Inhibits Hypertrophic Scar Formation by Inducing ROS-Mediated Apoptosis.Front Bioeng Biotechnol. 2022 May 24;10:892015. doi: 10.3389/fbioe.2022.892015. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35685086 Free PMC article.
References
-
- Yokoyama C, Wang X, Briggs MR, et al. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. - PubMed
-
- Rawson RB, Zelenski NG, Nijhawan D, et al. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell. 1997;1:47–57. - PubMed
-
- Sakai J, Rawson RB, Espenshade PJ, et al. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol Cell. 1998;2:505–514. - PubMed
-
- Briggs MR, Yokoyama C, Wang X, et al. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. I. Identification of the protein and delineation of its target nucleotide sequence. J Biol Chem. 1993;268:14490–14496. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
