Understanding the Binding Induced Folding of Intrinsically Disordered Proteins by Protein Engineering: Caveats and Pitfalls
- PMID: 32429036
- PMCID: PMC7279032
- DOI: 10.3390/ijms21103484
Understanding the Binding Induced Folding of Intrinsically Disordered Proteins by Protein Engineering: Caveats and Pitfalls
Abstract
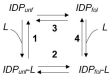
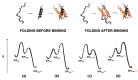
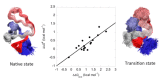
Many proteins lack a well-defined three-dimensional structure in isolation. These proteins, typically denoted as intrinsically disordered proteins (IDPs), may display a characteristic disorder-to-order transition when binding their physiological partner(s). From an experimental perspective, it is of great importance to establish the general grounds to understand how such folding processes may be explored. Here we discuss the caveats and the pitfalls arising when applying to IDPs one of the key techniques to characterize the folding of globular proteins, the Φ value analysis. This method is based on measurements of the free energy changes of transition and native states upon conservative, non-disrupting, mutations. On the basis of available data, we reinforce the validity of Φ value analysis in the study of IDPs and suggest future experiments to further validate this powerful experimental method.
Keywords: IDP; disorder-to order transition; folding kinetics; protein engineering; Φ value analysis.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
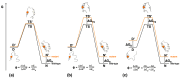
Similar articles
-
Folding and binding pathways of BH3-only proteins are encoded within their intrinsically disordered sequence, not templated by partner proteins.J Biol Chem. 2018 Jun 22;293(25):9718-9723. doi: 10.1074/jbc.RA118.002791. Epub 2018 May 1. J Biol Chem. 2018. PMID: 29716994 Free PMC article.
-
Templated folding of intrinsically disordered proteins.J Biol Chem. 2020 May 8;295(19):6586-6593. doi: 10.1074/jbc.REV120.012413. Epub 2020 Apr 6. J Biol Chem. 2020. PMID: 32253236 Free PMC article. Review.
-
Unveiling induced folding of intrinsically disordered proteins - Protein engineering, frustration and emerging themes.Curr Opin Struct Biol. 2022 Feb;72:153-160. doi: 10.1016/j.sbi.2021.11.004. Epub 2021 Dec 11. Curr Opin Struct Biol. 2022. PMID: 34902817 Review.
-
Analyzing the Folding and Binding Steps of an Intrinsically Disordered Protein by Protein Engineering.Biochemistry. 2017 Jul 25;56(29):3780-3786. doi: 10.1021/acs.biochem.7b00350. Epub 2017 Jul 11. Biochemistry. 2017. PMID: 28661120
-
Features of molecular recognition of intrinsically disordered proteins via coupled folding and binding.Protein Sci. 2019 Nov;28(11):1952-1965. doi: 10.1002/pro.3718. Epub 2019 Sep 4. Protein Sci. 2019. PMID: 31441158 Free PMC article. Review.
Cited by
-
Introducing intrinsic disorder reduces electrostatic steering in protein-protein interactions.Biophys J. 2021 Aug 3;120(15):2998-3007. doi: 10.1016/j.bpj.2021.06.021. Epub 2021 Jun 30. Biophys J. 2021. PMID: 34214536 Free PMC article.
-
Transient Co-expression of Membrane Protein Complexes in Mammalian Cells.Methods Mol Biol. 2024;2810:11-28. doi: 10.1007/978-1-0716-3878-1_2. Methods Mol Biol. 2024. PMID: 38926270
-
Liquid-Liquid Phase Separation and Protective Protein Aggregates in Bacteria.Molecules. 2023 Sep 12;28(18):6582. doi: 10.3390/molecules28186582. Molecules. 2023. PMID: 37764358 Free PMC article. Review.
-
Intrinsically Disordered Proteins: An Overview.Int J Mol Sci. 2022 Nov 14;23(22):14050. doi: 10.3390/ijms232214050. Int J Mol Sci. 2022. PMID: 36430530 Free PMC article. Review.
-
Non-synonymous variation and protein structure of candidate genes associated with selection in farm and wild populations of turbot (Scophthalmus maximus).Sci Rep. 2023 Feb 21;13(1):3019. doi: 10.1038/s41598-023-29826-z. Sci Rep. 2023. PMID: 36810752 Free PMC article.
References
-
- Adamski W., Salvi N., Maurin D., Magnat J., Milles S., Jensen M.R., Abyzov A., Moreau C.J., Blackledge M. A unified description of intrinsically disordered protein dynamics under physiological conditions using NMR spectroscopy. J. Am. Chem. Soc. 2019;141:17817–17829. doi: 10.1021/jacs.9b09002. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

