Atco, a yeast mitochondrial complex of Atp9 and Cox6, is an assembly intermediate of the ATP synthase
- PMID: 32413073
- PMCID: PMC7228087
- DOI: 10.1371/journal.pone.0233177
Atco, a yeast mitochondrial complex of Atp9 and Cox6, is an assembly intermediate of the ATP synthase
Abstract
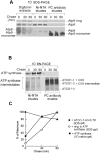
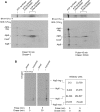
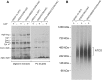
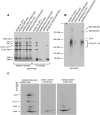
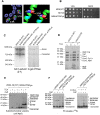
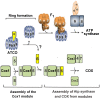
Mitochondrial oxidative phosphorylation (oxphos) is the process by which the ATP synthase conserves the energy released during the oxidation of different nutrients as ATP. The yeast ATP synthase consists of three assembly modules, one of which is a ring consisting of 10 copies of the Atp9 subunit. We previously reported the existence in yeast mitochondria of high molecular weight complexes composed of mitochondrially encoded Atp9 and of Cox6, an imported structural subunit of cytochrome oxidase (COX). Pulse-chase experiments indicated a correlation between the loss of newly translated Atp9 complexed to Cox6 and an increase of newly formed Atp9 ring, but did not exclude the possibility of an alternate source of Atp9 for ring formation. Here we have extended studies on the functions and structure of this complex, referred to as Atco. We show that Atco is the exclusive source of Atp9 for the ATP synthase assembly. Pulse-chase experiments show that newly translated Atp9, present in Atco, is converted to a ring, which is incorporated into the ATP synthase with kinetics characteristic of a precursor-product relationship. Even though Atco does not contain the ring form of Atp9, cross-linking experiments indicate that it is oligomeric and that the inter-subunit interactions are similar to those of the bona fide ring. We propose that, by providing Atp9 for biogenesis of ATP synthase, Atco complexes free Cox6 for assembly of COX. This suggests that Atco complexes may play a role in coordinating assembly and maintaining proper stoichiometry of the two oxphos enzymes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Allotopic expression of COX6 elucidates Atco-driven co-assembly of cytochrome oxidase and ATP synthase.Life Sci Alliance. 2023 Aug 21;6(11):e202301965. doi: 10.26508/lsa.202301965. Print 2023 Nov. Life Sci Alliance. 2023. PMID: 37604582 Free PMC article.
-
Oxa1 directly interacts with Atp9 and mediates its assembly into the mitochondrial F1Fo-ATP synthase complex.Mol Biol Cell. 2007 May;18(5):1897-908. doi: 10.1091/mbc.e06-10-0925. Epub 2007 Mar 7. Mol Biol Cell. 2007. PMID: 17344477 Free PMC article.
-
Modular assembly of yeast mitochondrial ATP synthase and cytochrome oxidase.Biol Chem. 2020 May 26;401(6-7):835-853. doi: 10.1515/hsz-2020-0112. Biol Chem. 2020. PMID: 32142477 Review.
-
Ribosome recycling defects modify the balance between the synthesis and assembly of specific subunits of the oxidative phosphorylation complexes in yeast mitochondria.Nucleic Acids Res. 2016 Jul 8;44(12):5785-97. doi: 10.1093/nar/gkw490. Epub 2016 Jun 1. Nucleic Acids Res. 2016. PMID: 27257059 Free PMC article.
-
Mechanisms of mitochondrial translational regulation.IUBMB Life. 2013 May;65(5):397-408. doi: 10.1002/iub.1156. Epub 2013 Apr 3. IUBMB Life. 2013. PMID: 23554047 Review.
Cited by
-
T1121G Point Mutation in the Mitochondrial Gene COX1 Suppresses a Null Mutation in ATP23 Required for the Assembly of Yeast Mitochondrial ATP Synthase.Int J Mol Sci. 2022 Feb 19;23(4):2327. doi: 10.3390/ijms23042327. Int J Mol Sci. 2022. PMID: 35216443 Free PMC article.
-
Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces cerevisiae.Life (Basel). 2020 Nov 23;10(11):304. doi: 10.3390/life10110304. Life (Basel). 2020. PMID: 33238568 Free PMC article. Review.
-
Allotopic expression of COX6 elucidates Atco-driven co-assembly of cytochrome oxidase and ATP synthase.Life Sci Alliance. 2023 Aug 21;6(11):e202301965. doi: 10.26508/lsa.202301965. Print 2023 Nov. Life Sci Alliance. 2023. PMID: 37604582 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

