Genotoxic stress triggers the activation of IRE1α-dependent RNA decay to modulate the DNA damage response
- PMID: 32409639
- PMCID: PMC7224204
- DOI: 10.1038/s41467-020-15694-y
Genotoxic stress triggers the activation of IRE1α-dependent RNA decay to modulate the DNA damage response
Abstract
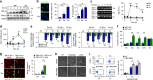
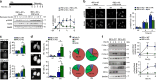
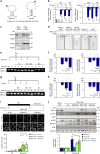
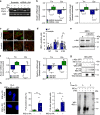
The molecular connections between homeostatic systems that maintain both genome integrity and proteostasis are poorly understood. Here we identify the selective activation of the unfolded protein response transducer IRE1α under genotoxic stress to modulate repair programs and sustain cell survival. DNA damage engages IRE1α signaling in the absence of an endoplasmic reticulum (ER) stress signature, leading to the exclusive activation of regulated IRE1α-dependent decay (RIDD) without activating its canonical output mediated by the transcription factor XBP1. IRE1α endoribonuclease activity controls the stability of mRNAs involved in the DNA damage response, impacting DNA repair, cell cycle arrest and apoptosis. The activation of the c-Abl kinase by DNA damage triggers the oligomerization of IRE1α to catalyze RIDD. The protective role of IRE1α under genotoxic stress is conserved in fly and mouse. Altogether, our results uncover an important intersection between the molecular pathways that sustain genome stability and proteostasis.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Peptides derived from the bifunctional kinase/RNase enzyme IRE1α modulate IRE1α activity and protect cells from endoplasmic reticulum stress.FASEB J. 2011 Sep;25(9):3115-29. doi: 10.1096/fj.11-182931. Epub 2011 Jun 16. FASEB J. 2011. PMID: 21680894
-
IRE1α activation protects mice against acetaminophen-induced hepatotoxicity.J Exp Med. 2012 Feb 13;209(2):307-18. doi: 10.1084/jem.20111298. Epub 2012 Jan 30. J Exp Med. 2012. PMID: 22291093 Free PMC article.
-
ER stress and distinct outputs of the IRE1α RNase control proliferation and senescence in response to oncogenic Ras.Proc Natl Acad Sci U S A. 2017 Sep 12;114(37):9900-9905. doi: 10.1073/pnas.1701757114. Epub 2017 Aug 28. Proc Natl Acad Sci U S A. 2017. PMID: 28847931 Free PMC article.
-
The multiple roles of the unfolded protein response regulator IRE1α in cancer.Mol Carcinog. 2019 Sep;58(9):1623-1630. doi: 10.1002/mc.23031. Epub 2019 Apr 30. Mol Carcinog. 2019. PMID: 31041814 Free PMC article. Review.
-
The unknown face of IRE1α - Beyond ER stress.Eur J Cell Biol. 2018 Jun;97(5):359-368. doi: 10.1016/j.ejcb.2018.05.002. Epub 2018 May 4. Eur J Cell Biol. 2018. PMID: 29747876 Review.
Cited by
-
Endoplasmic Reticulum Stress Signaling and the Pathogenesis of Hepatocarcinoma.Int J Mol Sci. 2021 Feb 11;22(4):1799. doi: 10.3390/ijms22041799. Int J Mol Sci. 2021. PMID: 33670323 Free PMC article. Review.
-
Zinc Supplementation Enhances the Pro-Death Function of UPR in Lymphoma Cells Exposed to Radiation.Biology (Basel). 2022 Jan 13;11(1):132. doi: 10.3390/biology11010132. Biology (Basel). 2022. PMID: 35053130 Free PMC article.
-
Senescence: a double-edged sword in beta-cell health and failure?Front Endocrinol (Lausanne). 2023 May 9;14:1196460. doi: 10.3389/fendo.2023.1196460. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37229454 Free PMC article.
-
A Syx-RhoA-Dia1 signaling axis regulates cell cycle progression, DNA damage, and therapy resistance in glioblastoma.JCI Insight. 2023 Jul 10;8(13):e157491. doi: 10.1172/jci.insight.157491. JCI Insight. 2023. PMID: 37427593 Free PMC article.
-
Adipocyte IRE1α promotes PGC1α mRNA decay and restrains adaptive thermogenesis.Nat Metab. 2022 Sep;4(9):1166-1184. doi: 10.1038/s42255-022-00631-8. Epub 2022 Sep 19. Nat Metab. 2022. PMID: 36123394
References
-
- Chevet E, Hetz C, Samali A. Endoplasmic reticulum stress–activated cell reprogramming in oncogenesis. Cancer Discov. 2016;5:586–597. doi: 10.1158/2159-8290.CD-14-1490. - DOI - PubMed
-
- Hetz, C. & Papa, F. R. The unfolded protein response and cell fate control. Mol. Cell. 10.1016/j.molcel.2017.06.017 (2018). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

