Dissecting the phenotypic and functional heterogeneity of mouse inflammatory osteoclasts by the expression of Cx3cr1
- PMID: 32400390
- PMCID: PMC7220377
- DOI: 10.7554/eLife.54493
Dissecting the phenotypic and functional heterogeneity of mouse inflammatory osteoclasts by the expression of Cx3cr1
Abstract
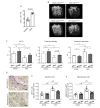
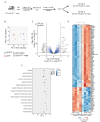
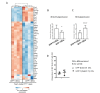
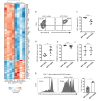
Bone destruction relies on interactions between bone and immune cells. Bone-resorbing osteoclasts (OCLs) were recently identified as innate immune cells activating T cells toward tolerance or inflammation. Thus, pathological bone destruction not only relies on increased osteoclast differentiation, but also on the presence of inflammatory OCLs (i-OCLs), part of which express Cx3cr1. Here, we investigated the contribution of mouse Cx3cr1+ and Cx3cr1neg i-OCLs to bone loss. We showed that Cx3cr1+ and Cx3cr1neg i-OCLs differ considerably in transcriptional and functional aspects. Cx3cr1neg i-OCLs have a high ability to resorb bone and activate inflammatory CD4+ T cells. Although Cx3cr1+ i-OCLs are associated with inflammation, they resorb less and have in vitro an immune-suppressive effect on Cx3cr1neg i-OCLs, mediated by PD-L1. Our results provide new insights into i-OCL heterogeneity. They also reveal that different i-OCL subsets may interact to regulate inflammation. This contributes to a better understanding and prevention of inflammatory bone destruction.
Keywords: bone destruction; cell biology; immunology; inflammation; mouse; osteoclast; osteoimmunology; osteoporosis.
Conflict of interest statement
MM, LI, TC, JH, MR, AB, CH, ID, FA, HG, AW, CB No competing interests declared
Figures









Similar articles
-
Immune Function and Diversity of Osteoclasts in Normal and Pathological Conditions.Front Immunol. 2019 Jun 19;10:1408. doi: 10.3389/fimmu.2019.01408. eCollection 2019. Front Immunol. 2019. PMID: 31275328 Free PMC article. Review.
-
Inflammatory Osteoclasts Prime TNFα-Producing CD4+ T Cells and Express CX3 CR1.J Bone Miner Res. 2016 Oct;31(10):1899-1908. doi: 10.1002/jbmr.2868. Epub 2016 Jun 30. J Bone Miner Res. 2016. PMID: 27161765
-
A Novel Reliable and Efficient Procedure for Purification of Mature Osteoclasts Allowing Functional Assays in Mouse Cells.Front Immunol. 2018 Nov 2;9:2567. doi: 10.3389/fimmu.2018.02567. eCollection 2018. Front Immunol. 2018. PMID: 30450105 Free PMC article.
-
Vascular expression of the chemokine CX3CL1 promotes osteoclast recruitment and exacerbates bone resorption in an irradiated murine model.Bone. 2014 Apr;61:91-101. doi: 10.1016/j.bone.2013.12.032. Epub 2014 Jan 5. Bone. 2014. PMID: 24401612
-
The origins and formation of bone-resorbing osteoclasts.Bone. 2022 Nov;164:116538. doi: 10.1016/j.bone.2022.116538. Epub 2022 Aug 23. Bone. 2022. PMID: 36028118 Review.
Cited by
-
Novel insights into the coupling of osteoclasts and resorption to bone formation.Semin Cell Dev Biol. 2022 Mar;123:4-13. doi: 10.1016/j.semcdb.2021.10.008. Epub 2021 Oct 30. Semin Cell Dev Biol. 2022. PMID: 34756783 Free PMC article. Review.
-
Interleukin-23 Regulates Inflammatory Osteoclastogenesis via Activation of CLEC5A(+) Osteoclast Precursors.Arthritis Rheumatol. 2023 Aug;75(8):1477-1489. doi: 10.1002/art.42478. Epub 2023 Jun 6. Arthritis Rheumatol. 2023. PMID: 36787107 Free PMC article.
-
Origin of Osteoclasts: Osteoclast Precursor Cells.J Bone Metab. 2023 May;30(2):127-140. doi: 10.11005/jbm.2023.30.2.127. Epub 2023 May 31. J Bone Metab. 2023. PMID: 37449346 Free PMC article.
-
The emerging role of osteoclasts in the treatment of bone metastases: rationale and recent clinical evidence.Front Oncol. 2024 Aug 1;14:1445025. doi: 10.3389/fonc.2024.1445025. eCollection 2024. Front Oncol. 2024. PMID: 39148909 Free PMC article. Review.
-
Inhibition of inflammatory osteoclasts accelerates callus remodeling in osteoporotic fractures by enhancing CGRP+TrkA+ signaling.Cell Death Differ. 2024 Dec;31(12):1695-1706. doi: 10.1038/s41418-024-01368-5. Epub 2024 Sep 2. Cell Death Differ. 2024. PMID: 39223264 Free PMC article.
References
-
- Ammari M, Presumey J, Ponsolles C, Roussignol G, Roubert C, Escriou V, Toupet K, Mausset-Bonnefont AL, Cren M, Robin M, Georgel P, Nehmar R, Taams L, Grün J, Grützkau A, Häupl T, Pers YM, Jorgensen C, Duroux-Richard I, Courties G, Apparailly F. Delivery of miR-146a to Ly6C high monocytes inhibits pathogenic bone erosion in inflammatory arthritis. Theranostics. 2018;8:5972–5985. doi: 10.7150/thno.29313. - DOI - PMC - PubMed
-
- Arai F, Miyamoto T, Ohneda O, Inada T, Sudo T, Brasel K, Miyata T, Anderson DM, Suda T. Commitment and differentiation of osteoclast precursor cells by the sequential expression of c-Fms and receptor activator of nuclear factor kappaB (RANK) receptors. The Journal of Experimental Medicine. 1999;190:1741–1754. doi: 10.1084/jem.190.12.1741. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

