Post-transcriptional regulation of several biological processes involved in latex production in Hevea brasiliensis
- PMID: 32391199
- PMCID: PMC7195832
- DOI: 10.7717/peerj.8932
Post-transcriptional regulation of several biological processes involved in latex production in Hevea brasiliensis
Abstract
Background: Small RNAs modulate plant gene expression at both the transcriptional and post-transcriptional level, mostly through the induction of either targeted DNA methylation or transcript cleavage, respectively. Small RNA networks are involved in specific plant developmental processes, in signaling pathways triggered by various abiotic stresses and in interactions between the plant and viral and non-viral pathogens. They are also involved in silencing maintenance of transposable elements and endogenous viral elements. Alteration in small RNA production in response to various environmental stresses can affect all the above-mentioned processes. In rubber trees, changes observed in small RNA populations in response to trees affected by tapping panel dryness, in comparison to healthy ones, suggest a shift from a transcriptional to a post-transcriptional regulatory pathway. This is the first attempt to characterise small RNAs involved in post-transcriptional silencing and their target transcripts in Hevea.
Methods: Genes producing microRNAs (MIR genes) and loci producing trans-activated small interfering RNA (ta-siRNA) were identified in the clone PB 260 re-sequenced genome. Degradome libraries were constructed with a pool of total RNA from six different Hevea tissues in stressed and non-stressed plants. The analysis of cleaved RNA data, associated with genomics and transcriptomics data, led to the identification of transcripts that are affected by 20-22 nt small RNA-mediated post-transcriptional regulation. A detailed analysis was carried out on gene families related to latex production and in response to growth regulators.
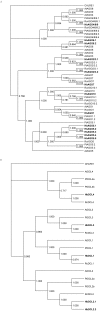
Results: Compared to other tissues, latex cells had a higher proportion of transcript cleavage activity mediated by miRNAs and ta-siRNAs. Post-transcriptional regulation was also observed at each step of the natural rubber biosynthesis pathway. Among the genes involved in the miRNA biogenesis pathway, our analyses showed that all of them are expressed in latex. Using phylogenetic analyses, we show that both the Argonaute and Dicer-like gene families recently underwent expansion. Overall, our study underlines the fact that important biological pathways, including hormonal signalling and rubber biosynthesis, are subject to post-transcriptional silencing in laticifers.
Keywords: Abiotic stress; Crop epigenomics; Degradome; Latex; Rubber tree; Small RNA; miRNA.
©2020 Leclercq et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


Similar articles
-
Structural and Functional Annotation of Transposable Elements Revealed a Potential Regulation of Genes Involved in Rubber Biosynthesis by TE-Derived siRNA Interference in Hevea brasiliensis.Int J Mol Sci. 2020 Jun 13;21(12):4220. doi: 10.3390/ijms21124220. Int J Mol Sci. 2020. PMID: 32545790 Free PMC article.
-
The small RNA profile in latex from Hevea brasiliensis trees is affected by tapping panel dryness.Tree Physiol. 2013 Oct;33(10):1084-98. doi: 10.1093/treephys/tpt076. Epub 2013 Nov 10. Tree Physiol. 2013. PMID: 24218245
-
Molecular characterization and expression analysis of the small GTPase ROP members expressed in laticifers of the rubber tree (Hevea brasiliensis).Plant Physiol Biochem. 2014 Jan;74:193-204. doi: 10.1016/j.plaphy.2013.11.013. Epub 2013 Nov 20. Plant Physiol Biochem. 2014. PMID: 24308989
-
Genomic technologies for Hevea breeding.Adv Genet. 2019;104:1-73. doi: 10.1016/bs.adgen.2019.04.001. Epub 2019 Jun 3. Adv Genet. 2019. PMID: 31200808 Review.
-
Tuning Beforehand: A Foresight on RNA Interference (RNAi) and In Vitro-Derived dsRNAs to Enhance Crop Resilience to Biotic and Abiotic Stresses.Int J Mol Sci. 2021 Jul 19;22(14):7687. doi: 10.3390/ijms22147687. Int J Mol Sci. 2021. PMID: 34299307 Free PMC article. Review.
Cited by
-
Downregulation of HbFPS1 affects rubber biosynthesis of Hevea brasiliensis suffering from tapping panel dryness.Plant J. 2023 Feb;113(3):504-520. doi: 10.1111/tpj.16063. Epub 2023 Jan 9. Plant J. 2023. PMID: 36524729 Free PMC article.
-
An Epigenetic Alphabet of Crop Adaptation to Climate Change.Front Genet. 2022 Feb 16;13:818727. doi: 10.3389/fgene.2022.818727. eCollection 2022. Front Genet. 2022. PMID: 35251130 Free PMC article. Review.
-
Structural and Functional Annotation of Transposable Elements Revealed a Potential Regulation of Genes Involved in Rubber Biosynthesis by TE-Derived siRNA Interference in Hevea brasiliensis.Int J Mol Sci. 2020 Jun 13;21(12):4220. doi: 10.3390/ijms21124220. Int J Mol Sci. 2020. PMID: 32545790 Free PMC article.
-
Genetic analysis of agronomic and physiological traits associated with latex yield revealed complex genetic bases in Hevea brasiliensis.Heliyon. 2024 Jun 22;10(13):e33421. doi: 10.1016/j.heliyon.2024.e33421. eCollection 2024 Jul 15. Heliyon. 2024. PMID: 39040337 Free PMC article.
-
Advancing image segmentation with DBO-Otsu: Addressing rubber tree diseases through enhanced threshold techniques.PLoS One. 2024 Mar 21;19(3):e0297284. doi: 10.1371/journal.pone.0297284. eCollection 2024. PLoS One. 2024. PMID: 38512907 Free PMC article.
References
-
- Ahmed F, Senthil-Kumar M, Lee S, Dai X, Mysore KS, Zhao PX. Comprehensive analysis of small RNA-seq data reveals that combination of miRNA with its isomiRs increase the accuracy of target prediction in Arabidopsis thaliana. RNA Biology. 2014;11:1414–1429. doi: 10.1080/15476286.2014.996474. - DOI - PMC - PubMed
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, Consortium GO. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. - DOI - PMC - PubMed
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

